library(tidyverse)
library(mdsr)
library(mosaicData)Stat. 651: ggplot2
ggplot2 examples
CIACounties
Make the base plot g and then add different layers on to it.
head(CIACountries) country pop area oil_prod gdp educ roadways net_users
1 Afghanistan 32564342 652230 0 1900 NA 0.06462444 >5%
2 Albania 3029278 28748 20510 11900 3.3 0.62613051 >35%
3 Algeria 39542166 2381741 1420000 14500 4.3 0.04771929 >15%
4 American Samoa 54343 199 0 13000 NA 1.21105528 <NA>
5 Andorra 85580 468 NA 37200 NA 0.68376068 >60%
6 Angola 19625353 1246700 1742000 7300 3.5 0.04125211 >15%# base plot g
g <- CIACountries %>% ggplot(aes(y = gdp, x = educ))
g + geom_point()Warning: Removed 64 rows containing missing values (geom_point).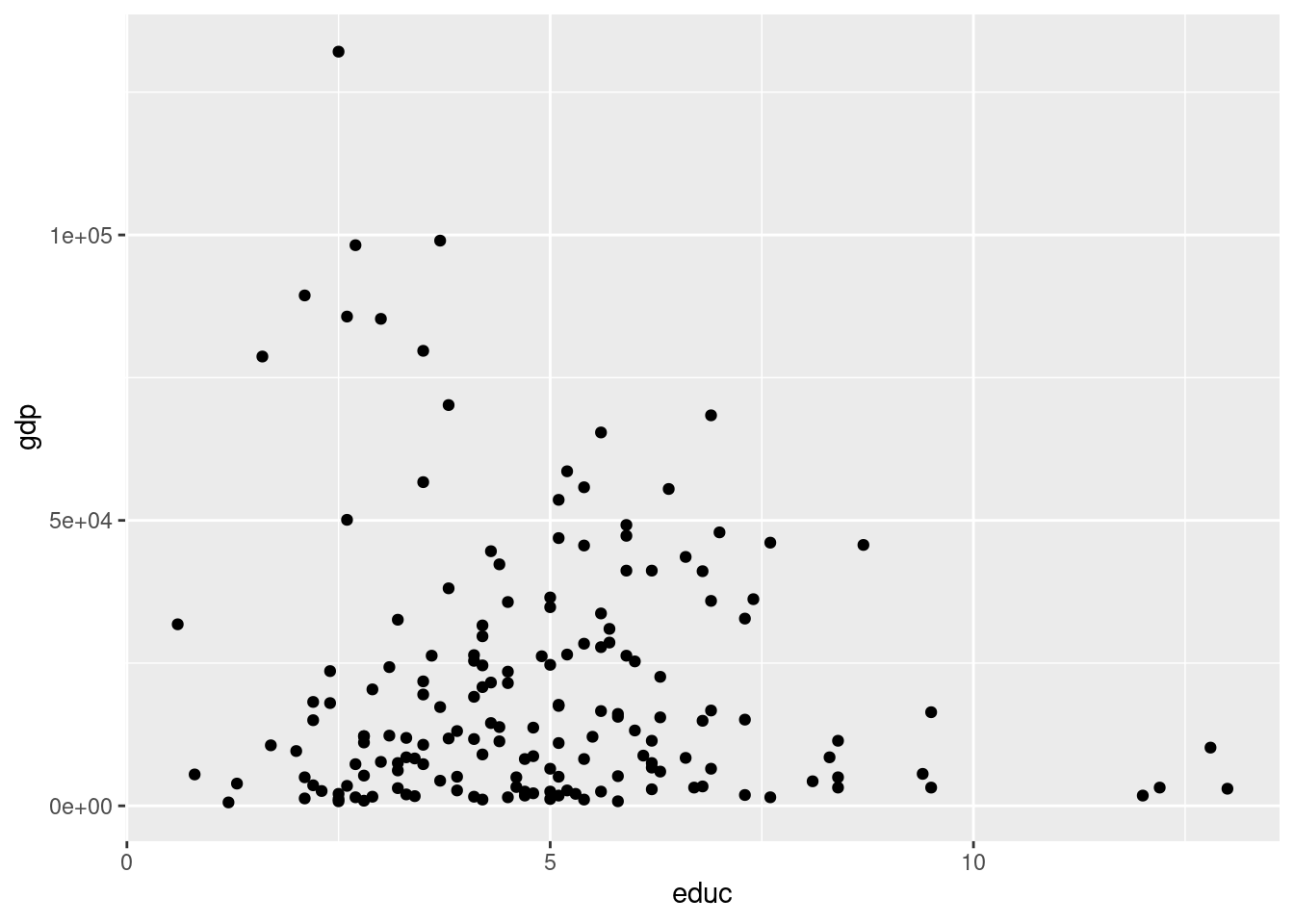
g + geom_point(size = 3)Warning: Removed 64 rows containing missing values (geom_point).
g + geom_point(aes(color = net_users), size = 3)Warning: Removed 64 rows containing missing values (geom_point).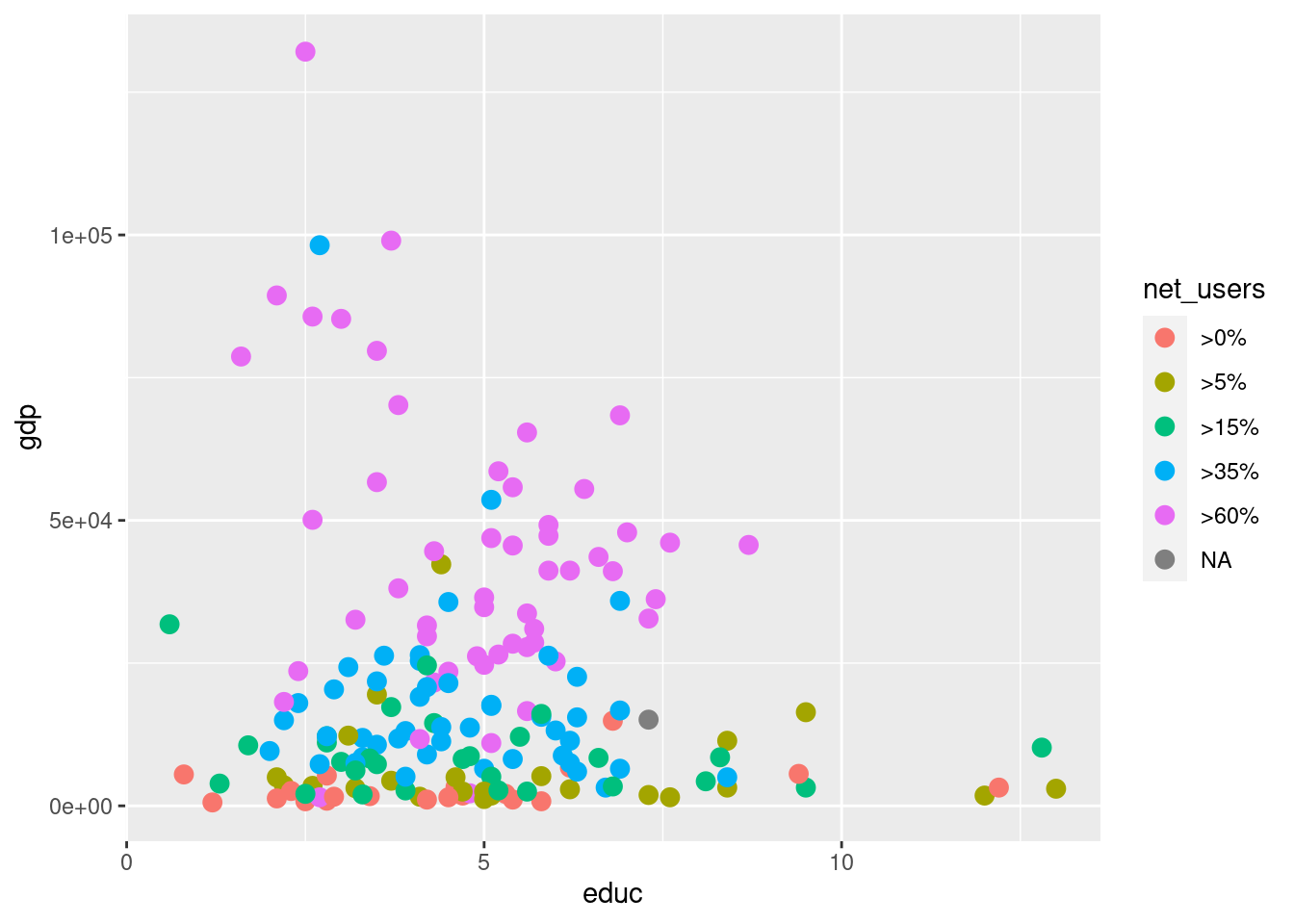
# no geom_point used for the next picture
g + geom_text( aes(label = country, color = net_users), size = 3 )Warning: Removed 64 rows containing missing values (geom_text).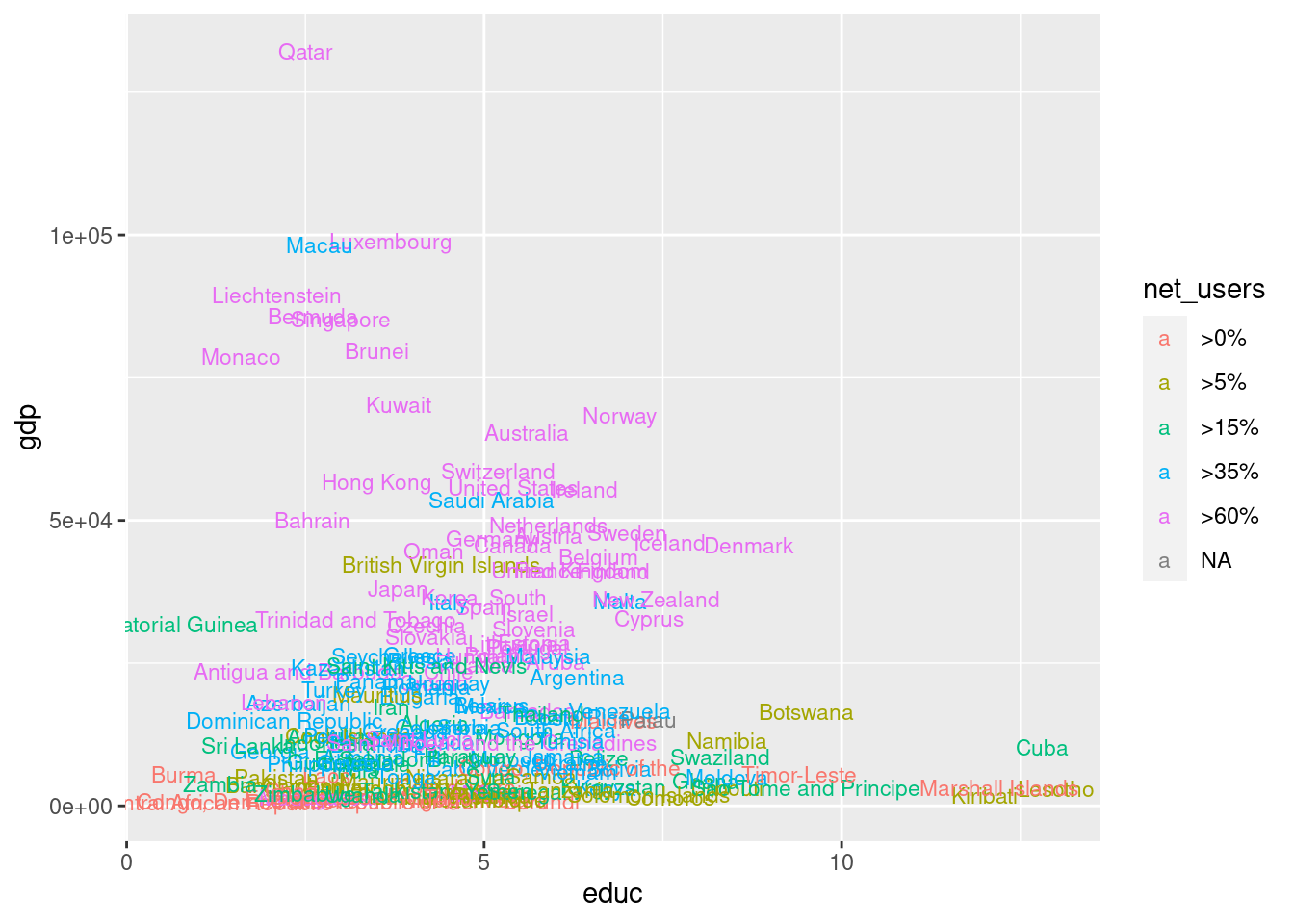
g + geom_point( aes(color = net_users, size = roadways) )Warning: Removed 66 rows containing missing values (geom_point).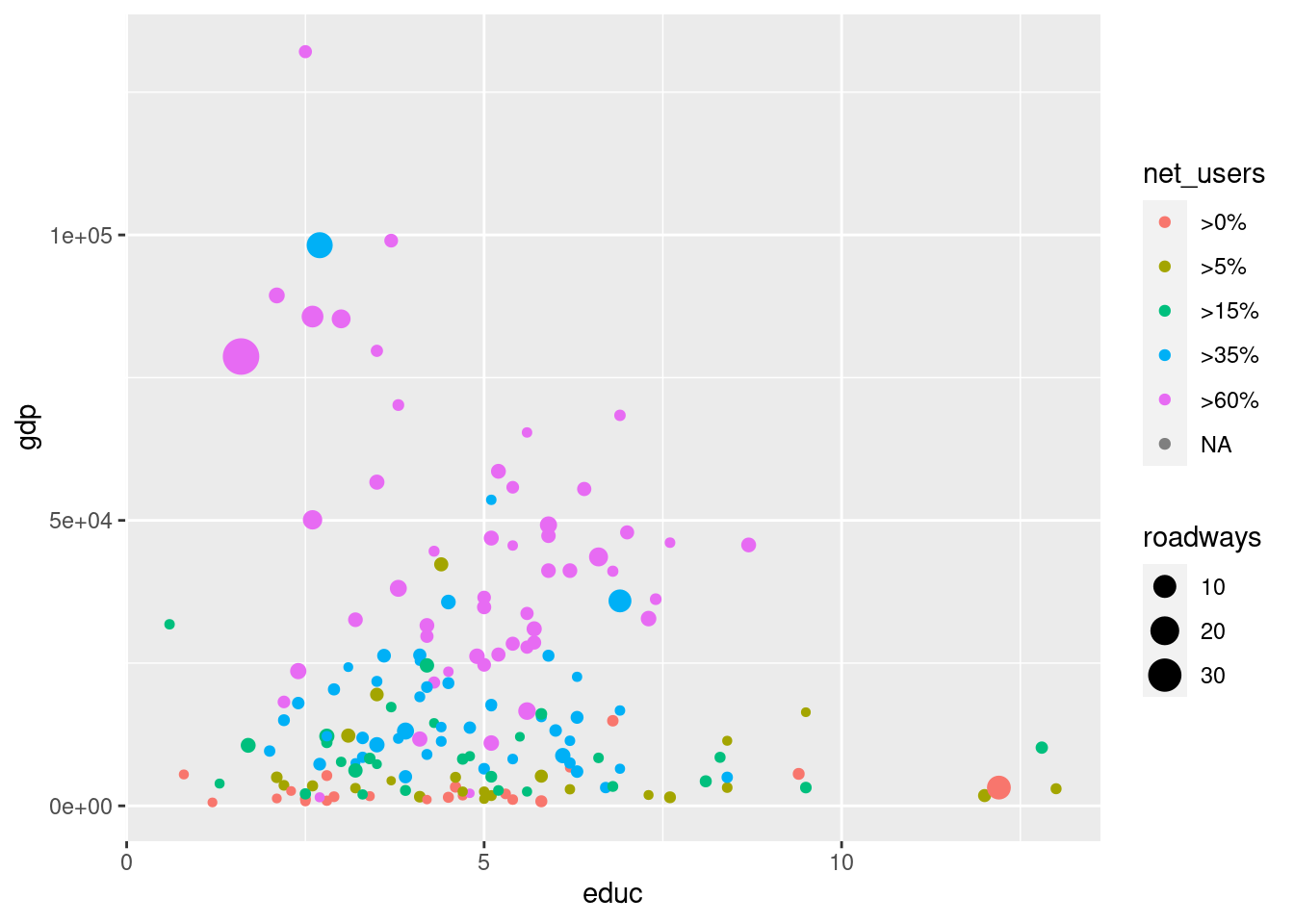
Change the scales
g + geom_point(aes(color = net_users, size = roadways)) +
coord_trans( y = "log10")Warning: Removed 66 rows containing missing values (geom_point).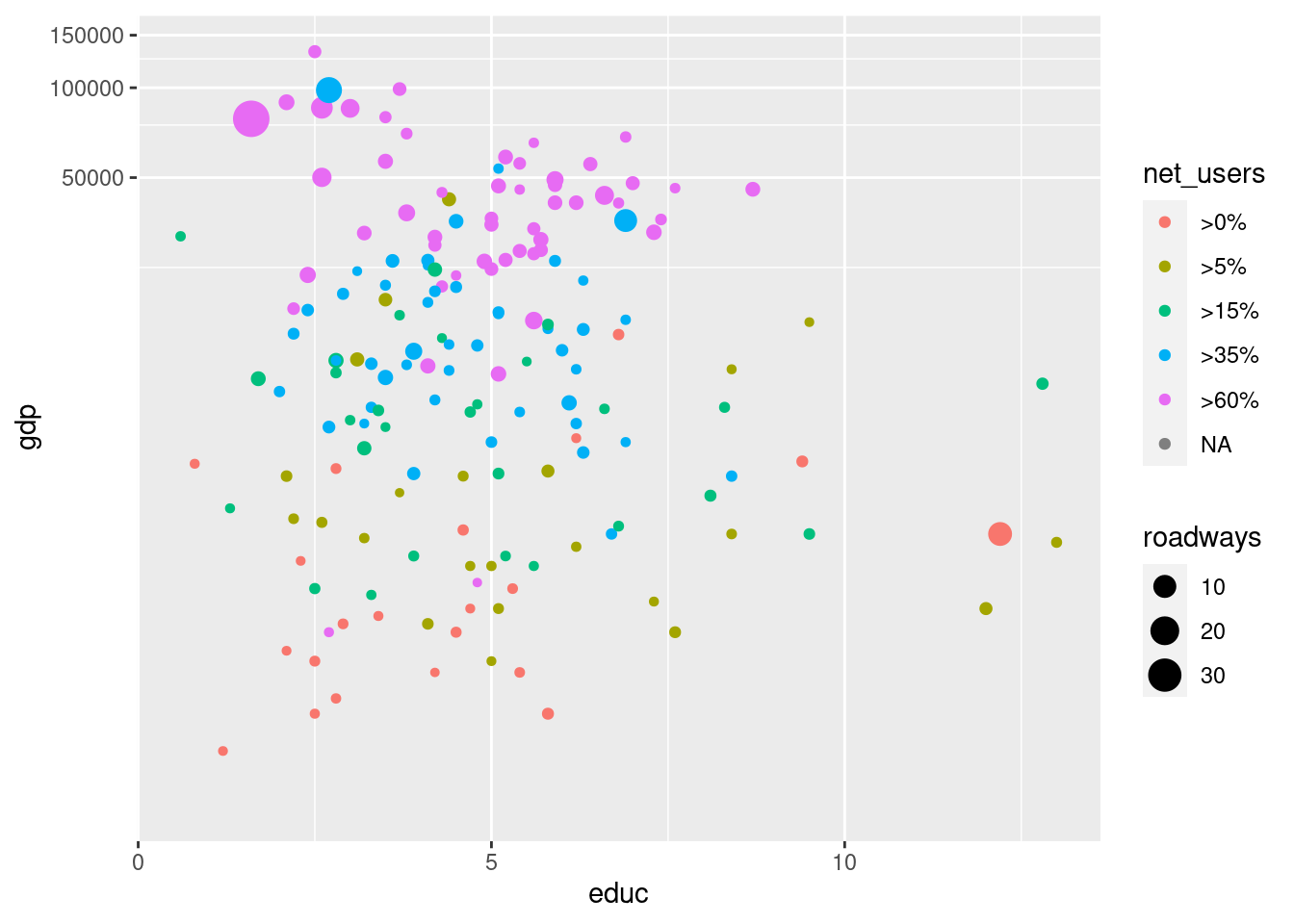
g + geom_point(aes(color = net_users, size = roadways)) +
scale_y_continuous(name = "Gross Domestic Product", trans = "log10")Warning: Removed 66 rows containing missing values (geom_point).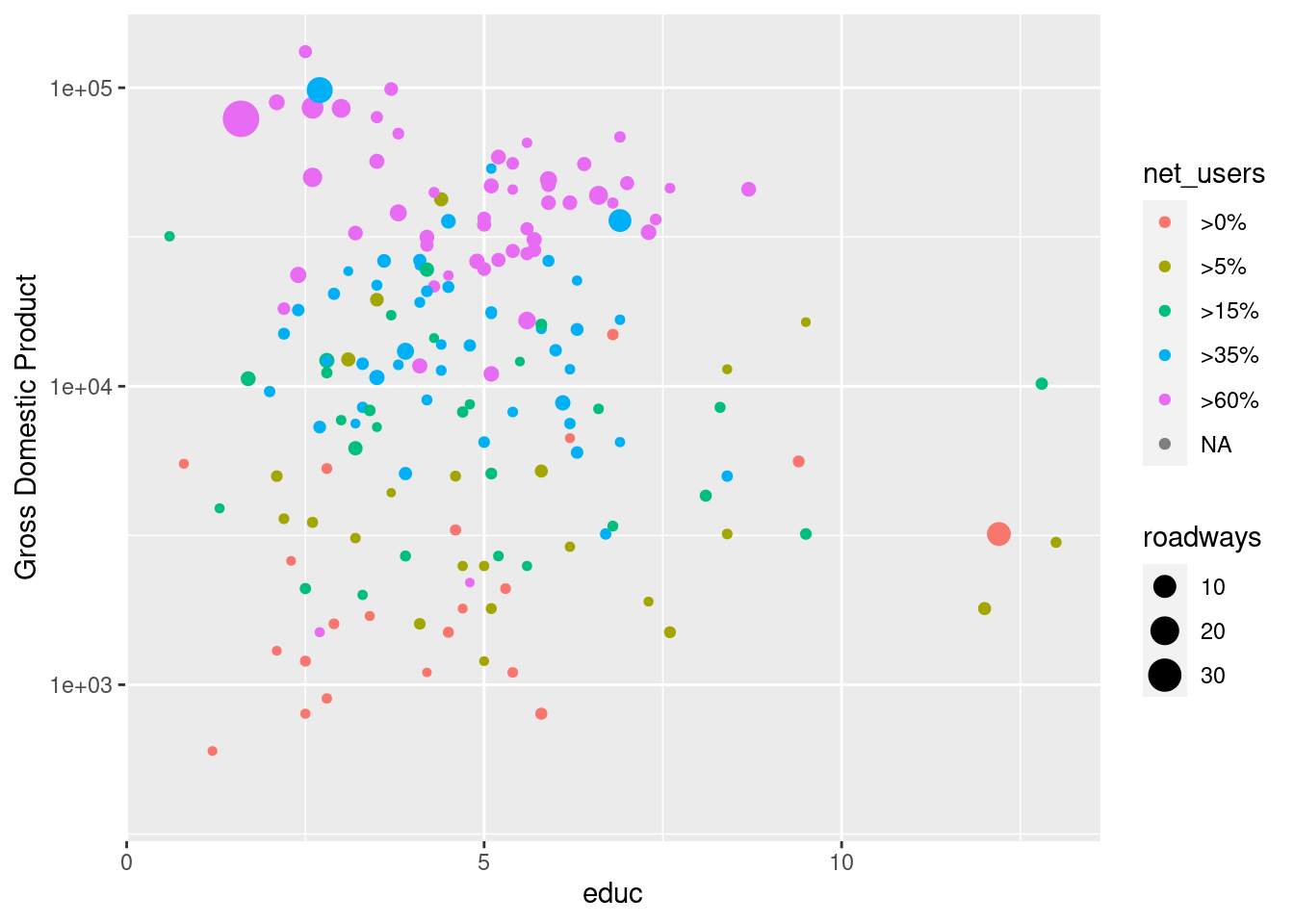
Faceting
g + geom_point(alpha = 0.9, aes(size = roadways)) +
coord_trans(y = "log10") +
facet_wrap( ~ net_users, nrow = 1) +
theme(legend.position = "top")Warning: Removed 66 rows containing missing values (geom_point).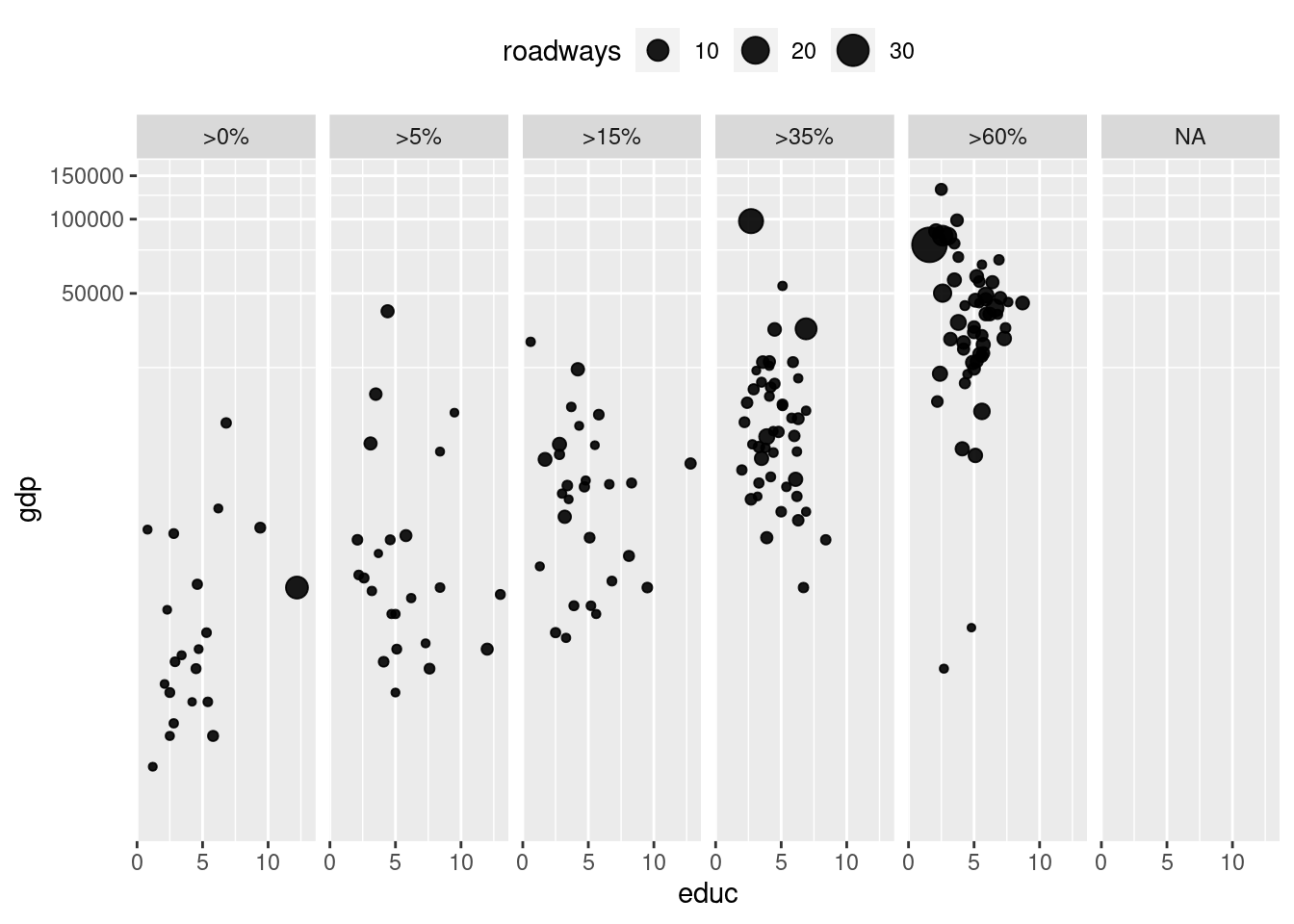
g + geom_point(alpha = 0.9, aes(size = roadways)) +
coord_trans(y = "log10") +
scale_y_continuous(name = "Gross Domestic Product", trans = "log10") +
facet_wrap( ~ net_users, nrow = 1) +
theme(legend.position = "top")Warning: Removed 66 rows containing missing values (geom_point).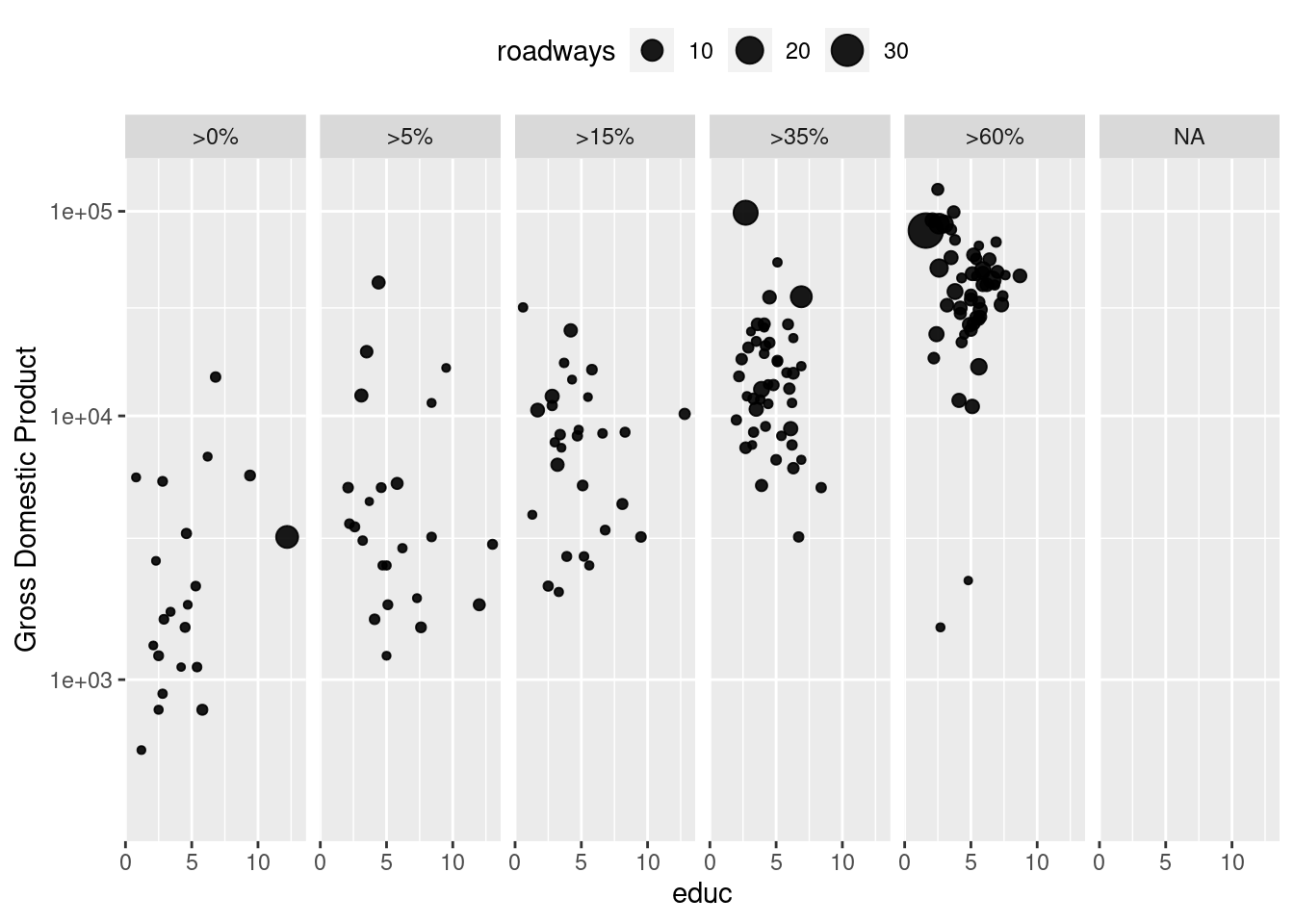
Export the data and try in Tableau
getwd()[1] "/home/esuess/classes/2022-2023/Fall 2022/Stat651/website/presentations/02_ggplot2"write_csv(CIACountries, "CIACountries.csv")MedicareCharges
Check out the MEPS website for more real data.
# head(MedicareCharges) # This now causes an error, remove the grouping.
? MedicareCharges
MedicareCharges <- ungroup(MedicareCharges)
head(MedicareCharges)# A tibble: 6 × 4
drg stateProvider num_charges mean_charge
<chr> <fct> <int> <dbl>
1 039 AK 1 34805.
2 039 AL 23 32044.
3 039 AR 16 27463.
4 039 AZ 24 33443.
5 039 CA 67 56095.
6 039 CO 10 35252.NJCharges <- MedicareCharges %>% filter(stateProvider == "NJ")
NJCharges# A tibble: 100 × 4
drg stateProvider num_charges mean_charge
<chr> <fct> <int> <dbl>
1 039 NJ 31 35104.
2 057 NJ 55 45692.
3 064 NJ 55 87042.
4 065 NJ 59 59576.
5 066 NJ 56 45819.
6 069 NJ 61 41917.
7 074 NJ 41 42993.
8 101 NJ 58 42314.
9 149 NJ 50 34916.
10 176 NJ 36 58941.
# … with 90 more rowsp <- NJCharges %>% ggplot(aes(y = mean_charge, x = reorder(drg, mean_charge))) +
geom_bar(fill = "grey", stat = "identity")
p
p <- p + ylab("Statewide Average Charges ($)") +
xlab("Medical Procedure (DRG)")
p
p <- p + theme(axis.text.x = element_text(angle = 90, hjust = 1))
p
Now add the overall data to the plot to compare with NJ.
p <- p + geom_point(data = MedicareCharges, size = 1, alpha = 0.3)
p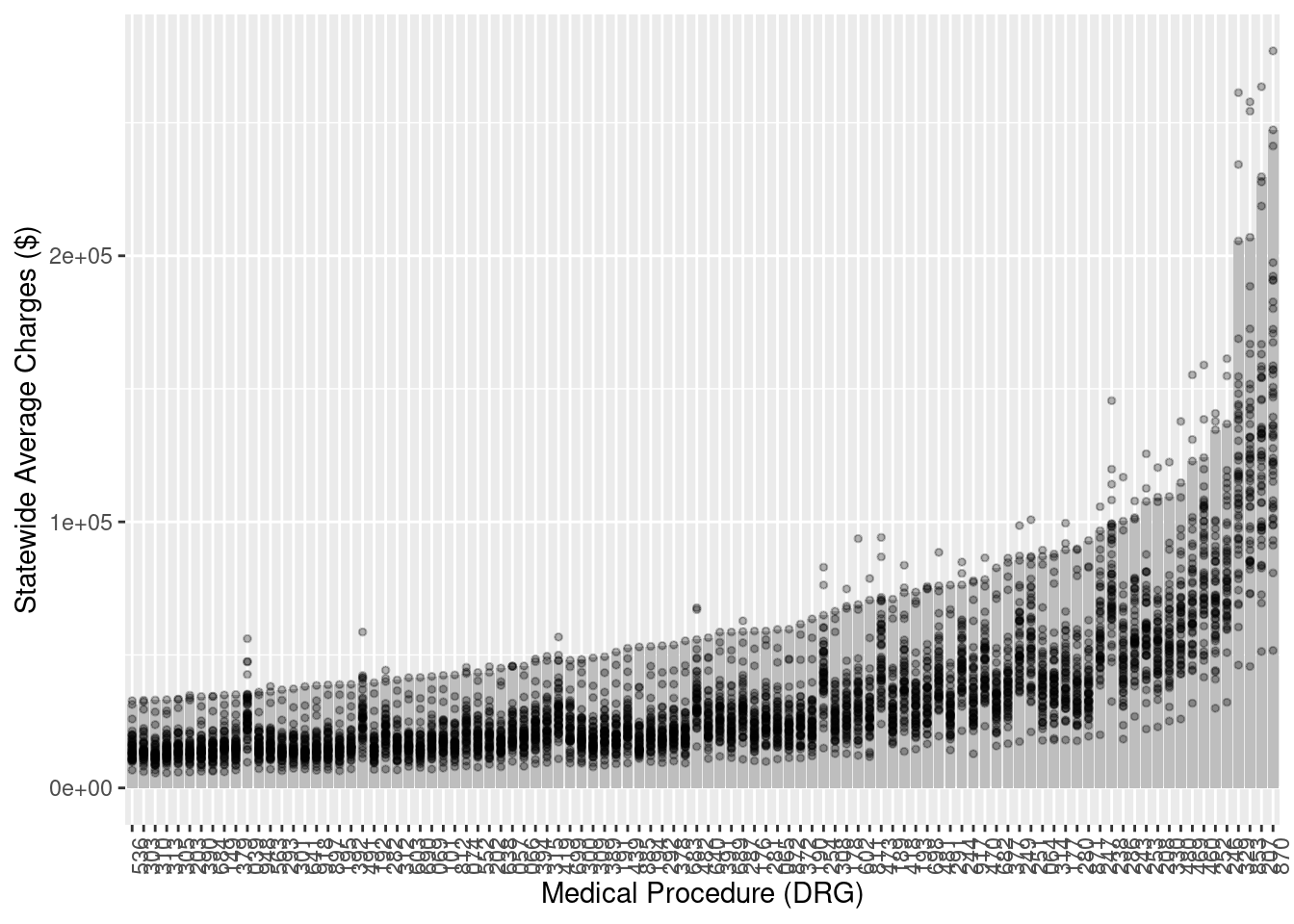
SAT
Here is the link to the College Board SAT website.
g <- SAT_2010 %>% ggplot(aes(x = math))
g + geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.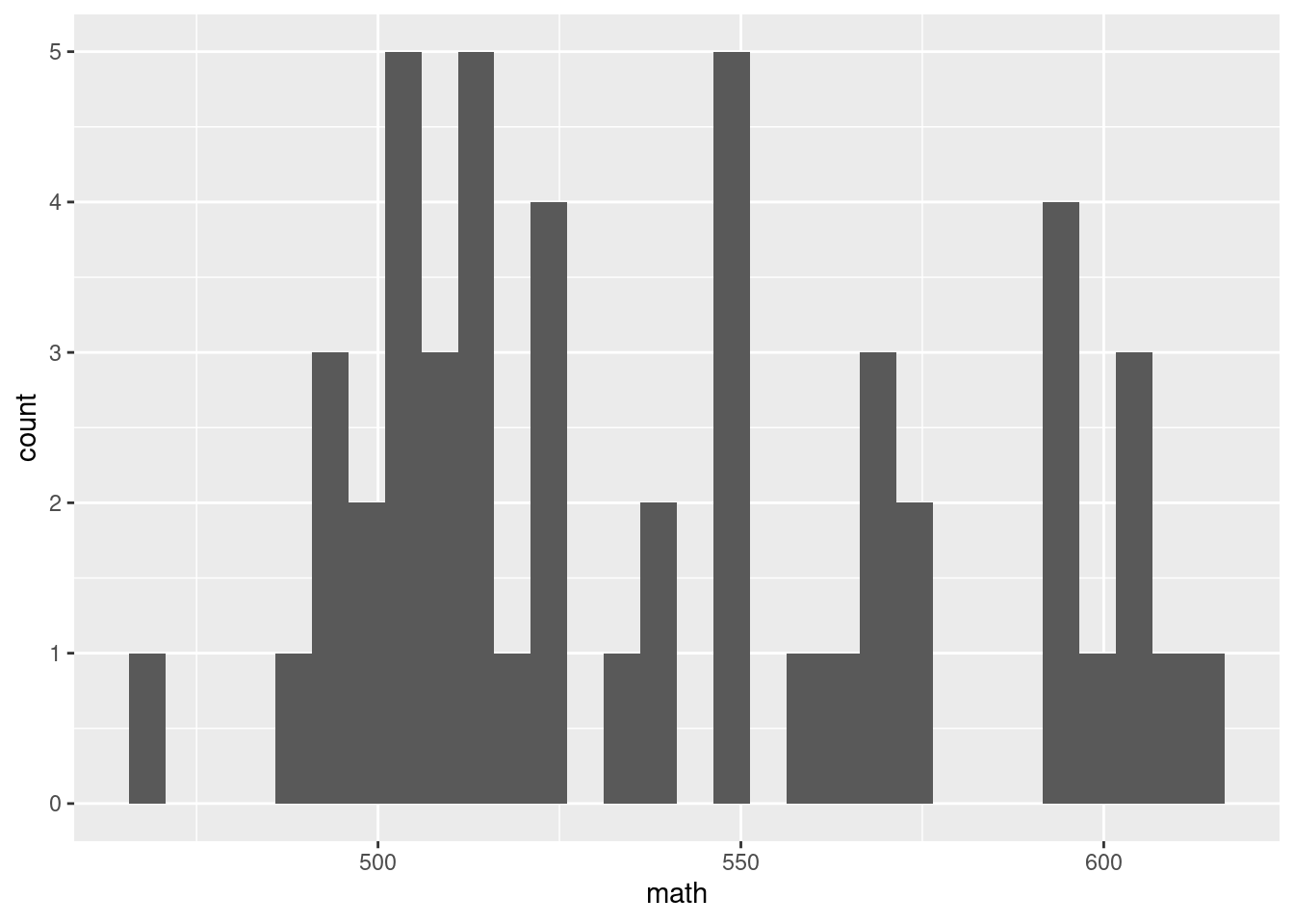
g + geom_histogram(binwidth = 10)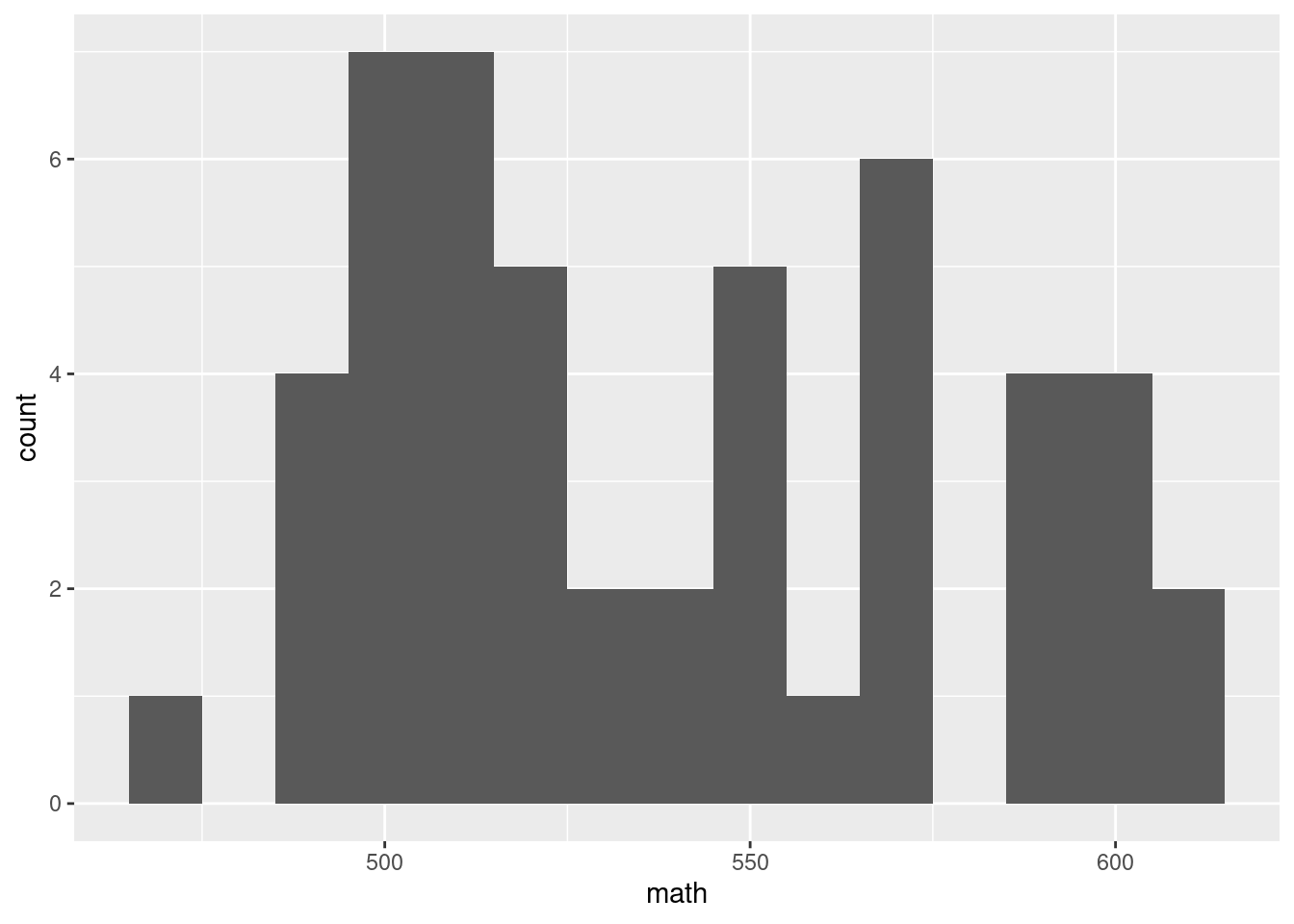
g + geom_density()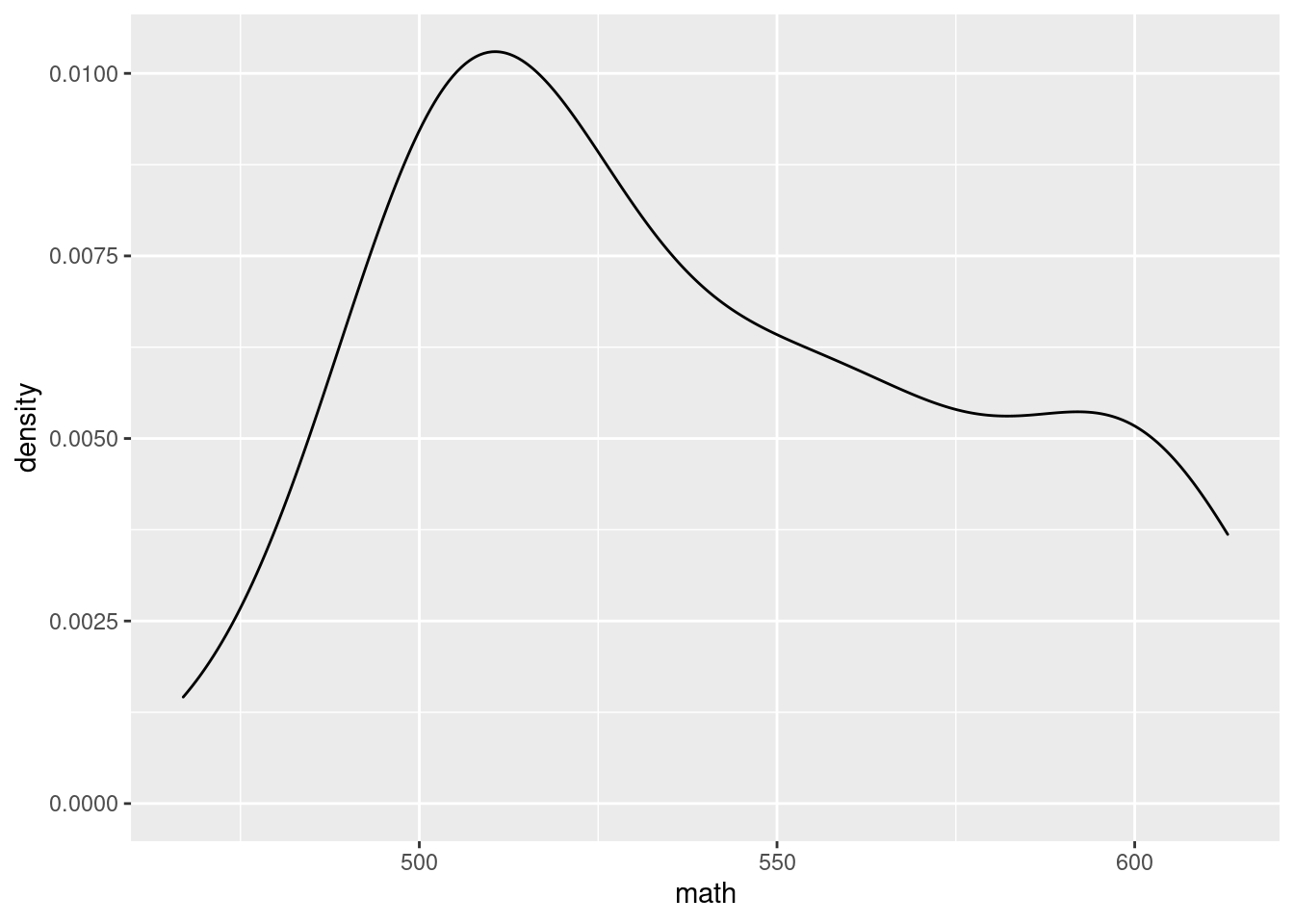
ggplot( data = head(SAT_2010, 10), aes( y = math, x = reorder(state, math) ) ) +
geom_bar(stat = "identity")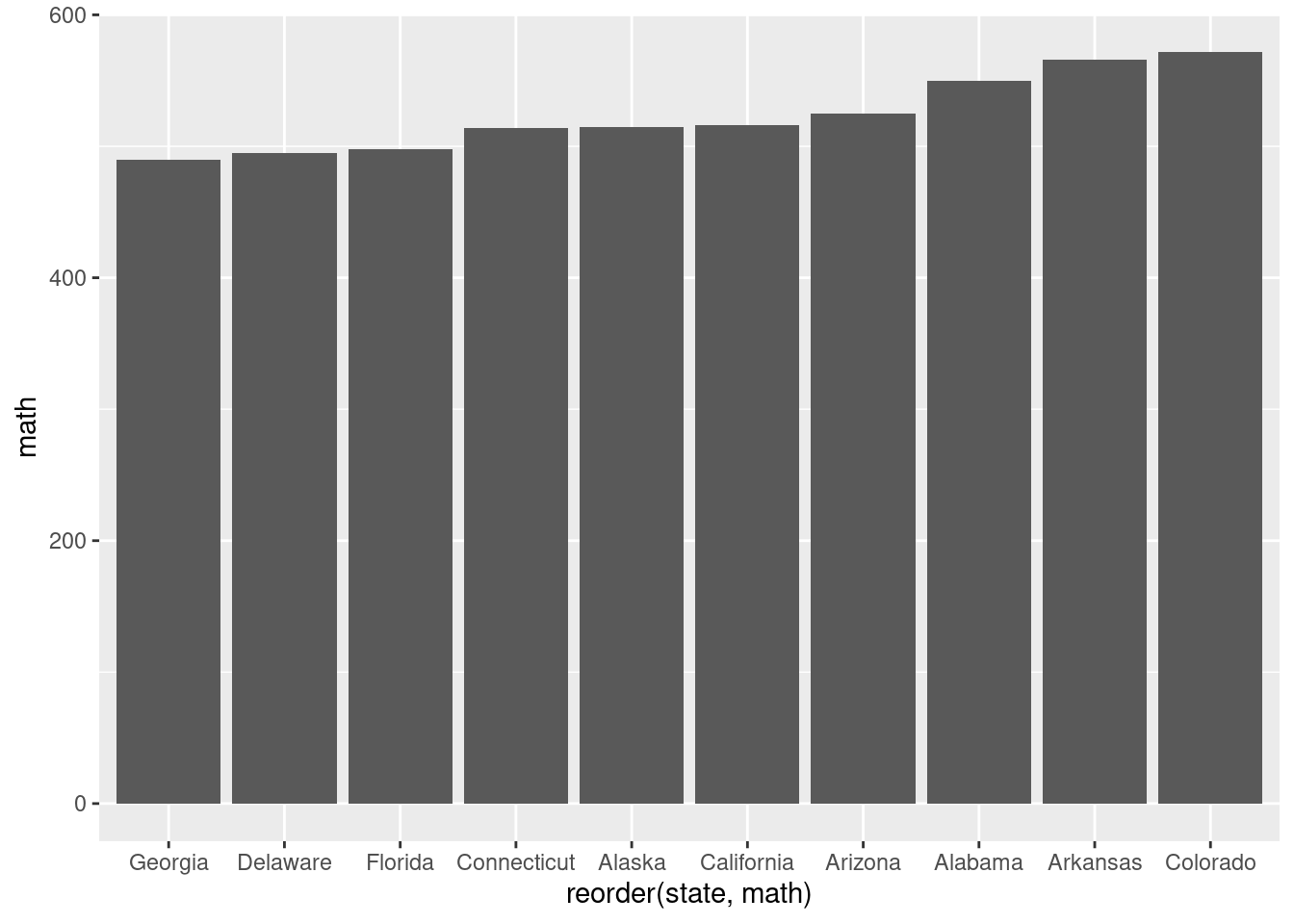
Scatterplot with tend lines
g <- SAT_2010 %>% ggplot(aes(x = expenditure, y = math)) +
geom_point()
g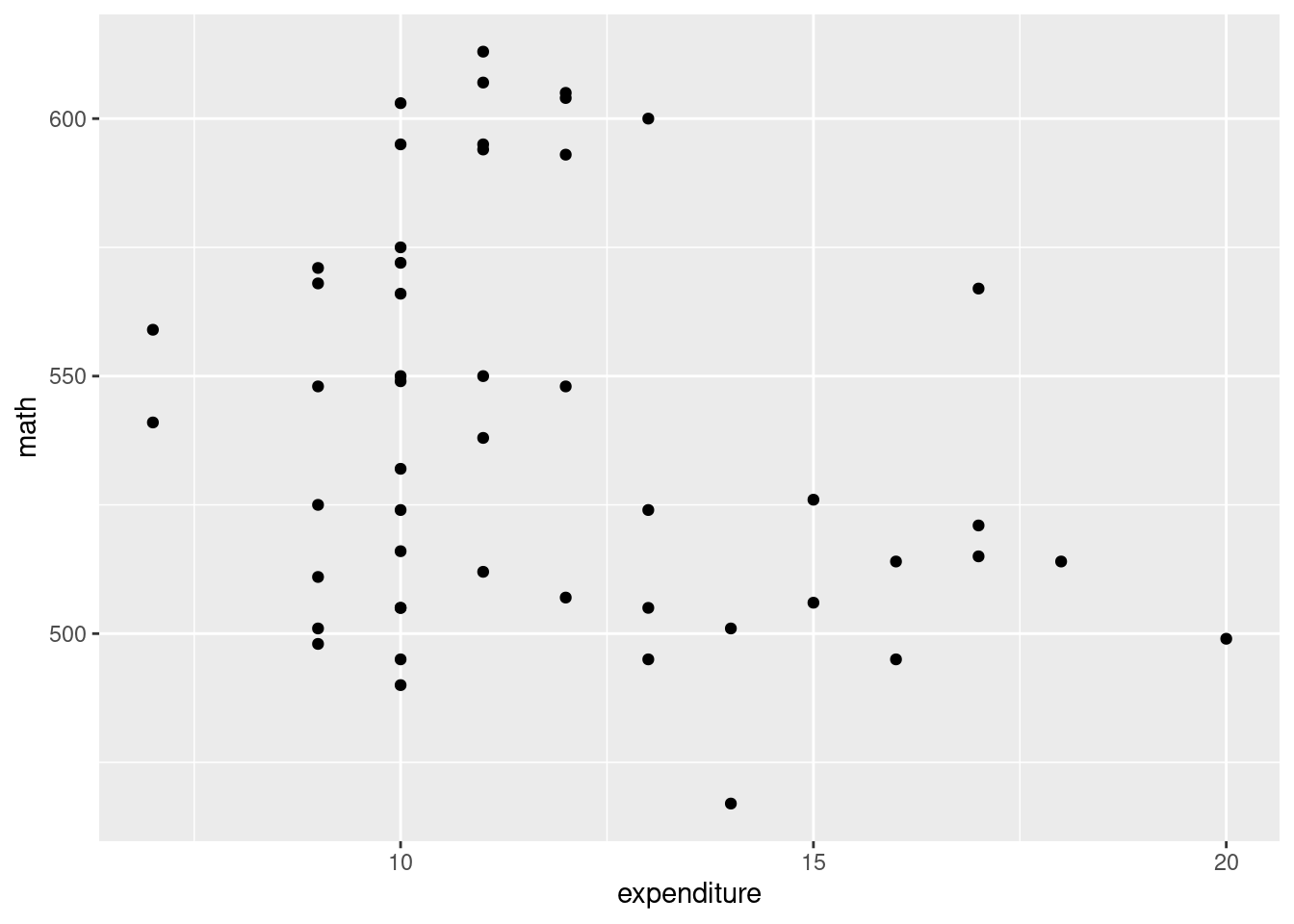
g <- g + geom_smooth(method="lm", se = 0) +
xlab("Average expenditure per student ($100)") +
ylab("Average score on math SAT")
g`geom_smooth()` using formula 'y ~ x'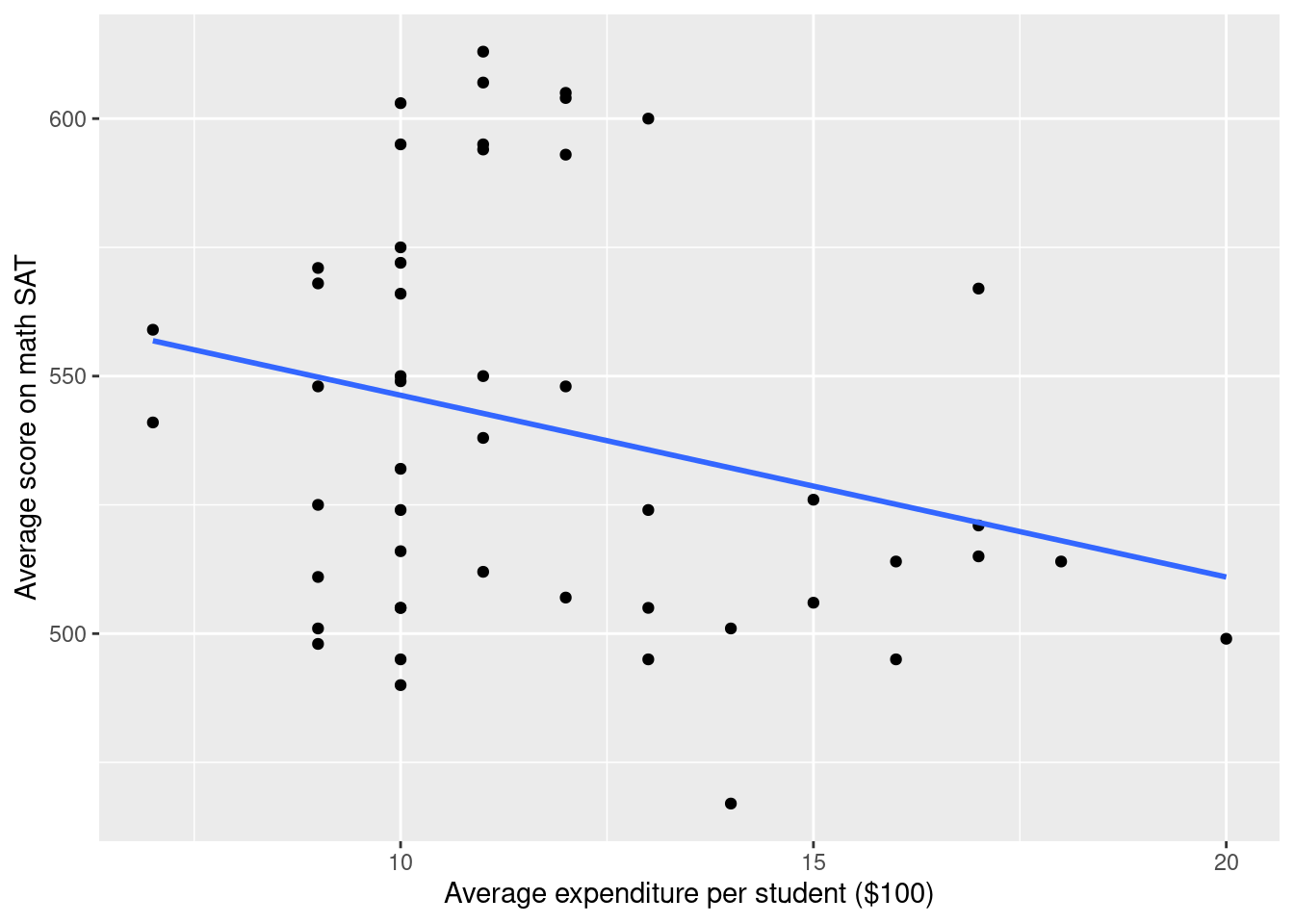
Add the trend line within groups representing rate of taking the test.
SAT_2010 <- SAT_2010 %>%
mutate(SAT_rate = cut(sat_pct, breaks = c(0, 30, 60, 100), labels = c("low", "medium", "high") ))
g <- g %+% SAT_2010
g`geom_smooth()` using formula 'y ~ x'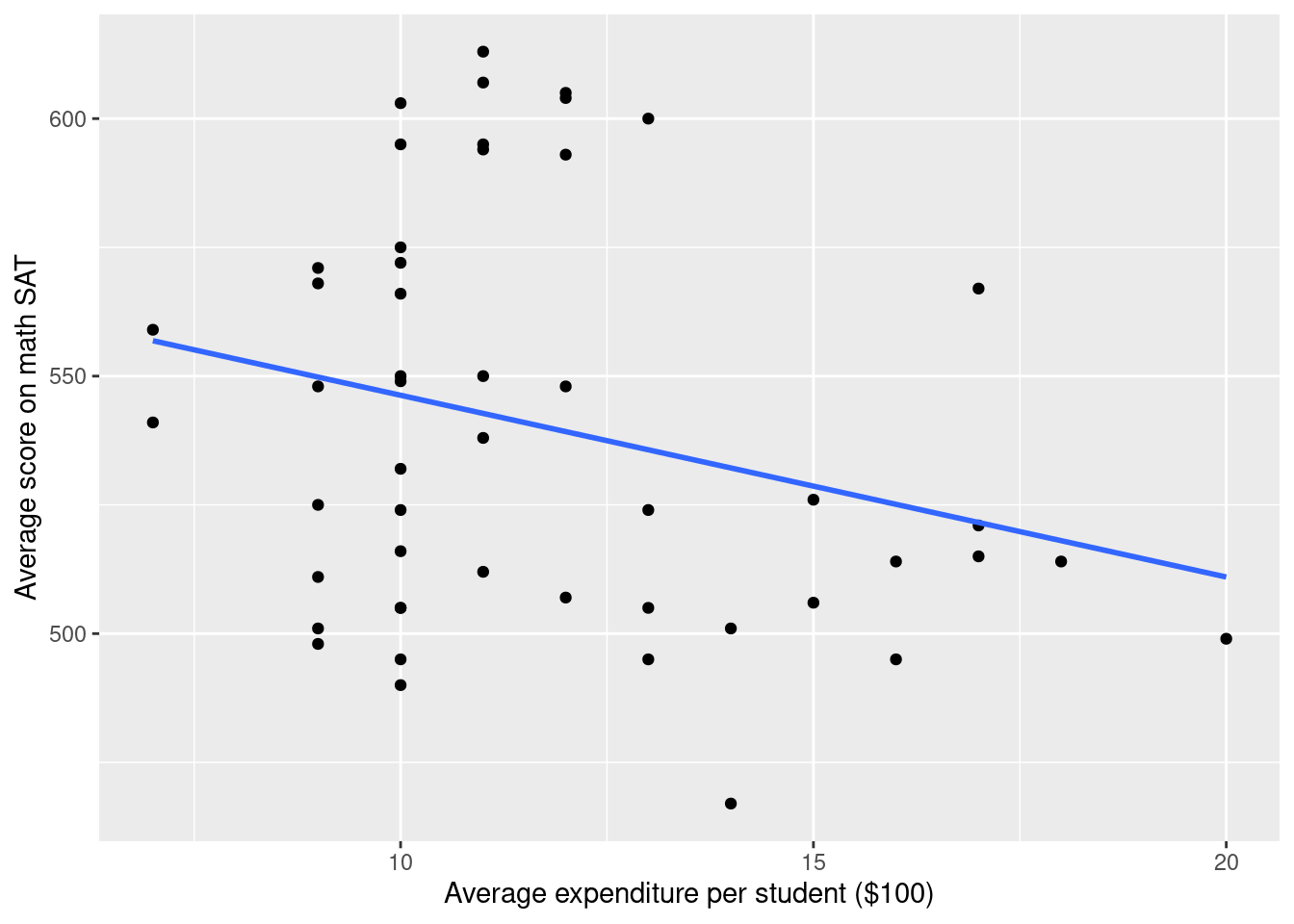
g + aes(color = SAT_rate)`geom_smooth()` using formula 'y ~ x'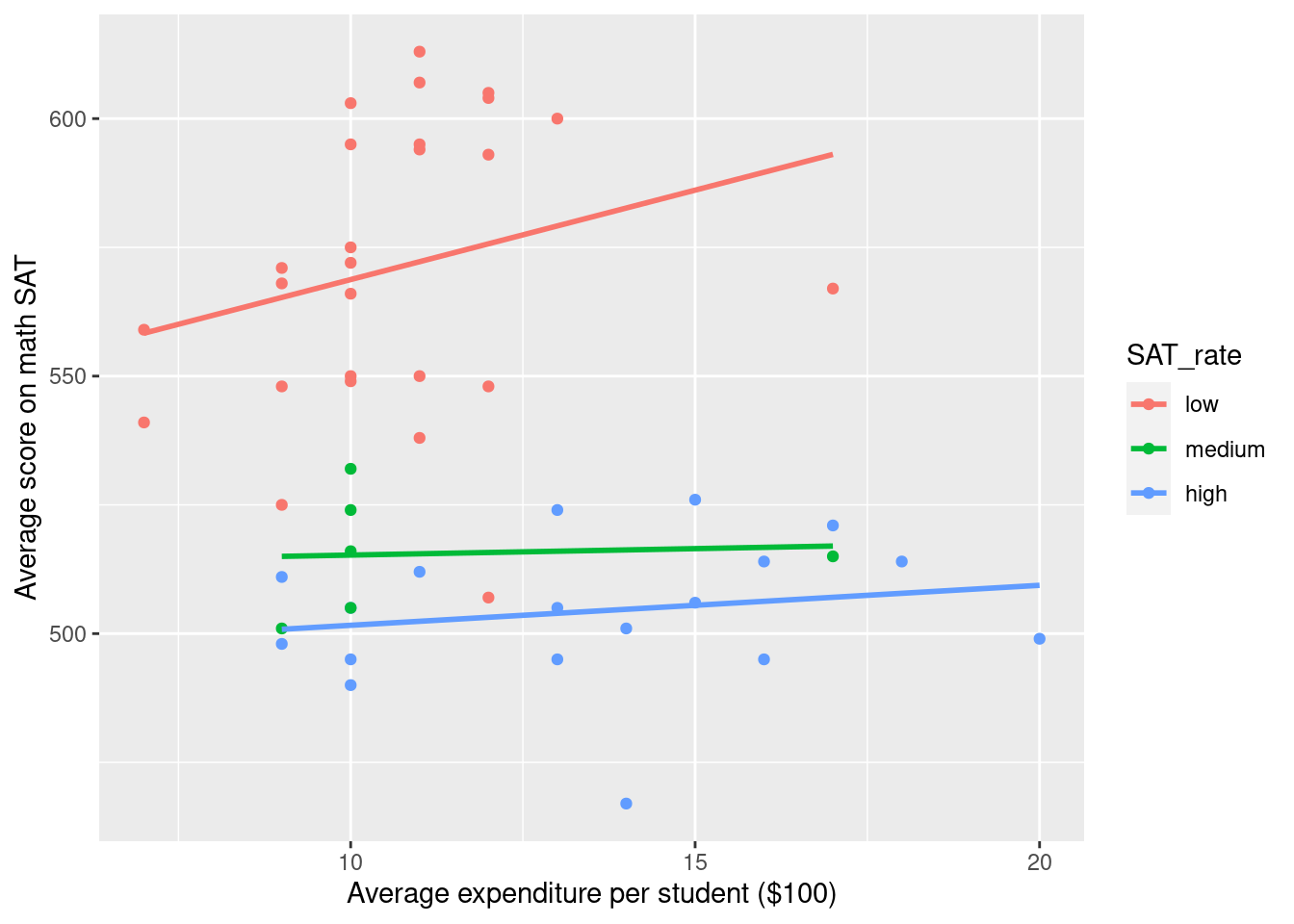
g +facet_wrap( ~ SAT_rate)`geom_smooth()` using formula 'y ~ x'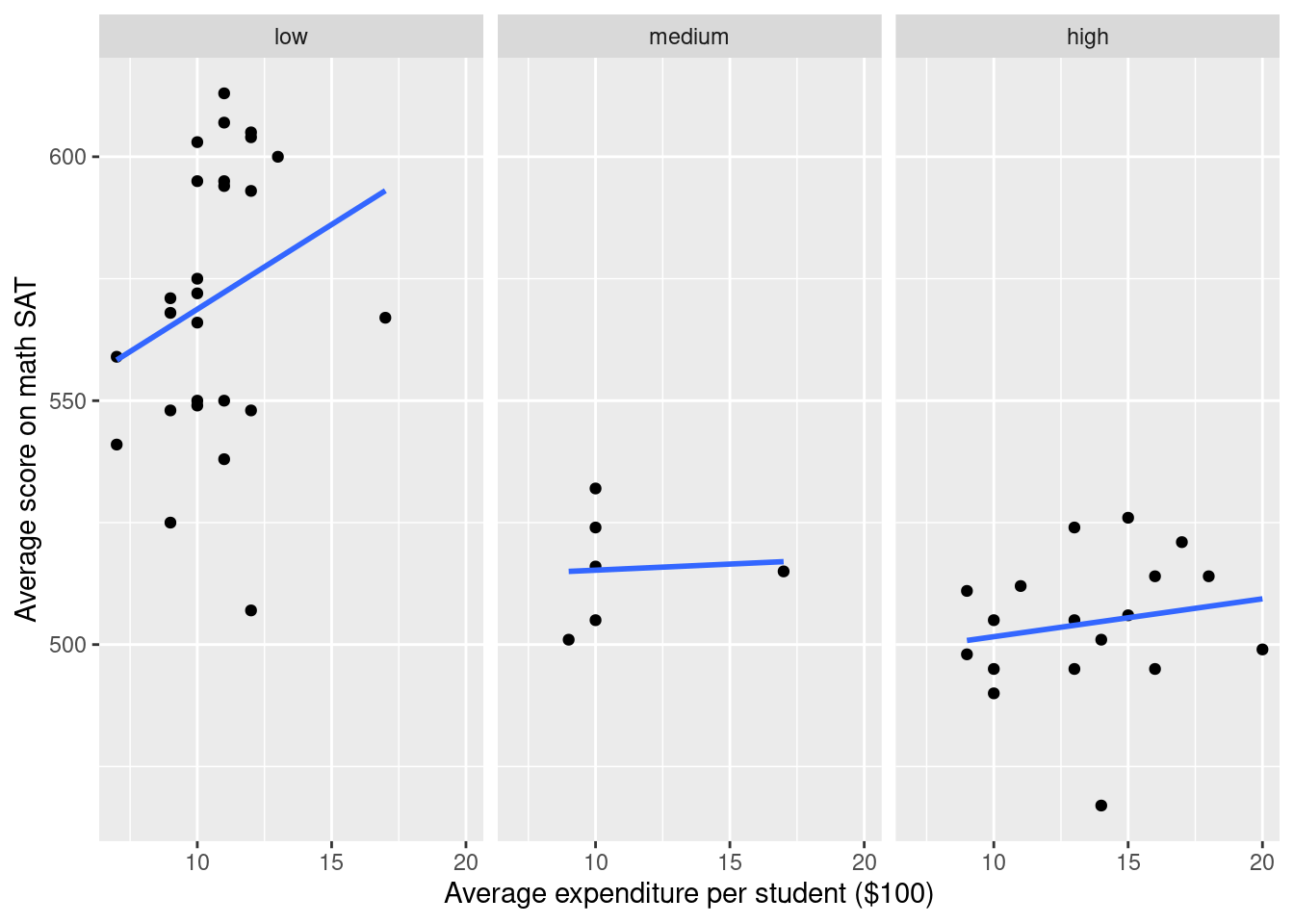
HELPPrct
Here is the link to the NSDUH website.
HELPrct %>% ggplot(aes(x = homeless)) +
geom_bar(aes(fill = substance), position = "fill") +
coord_flip()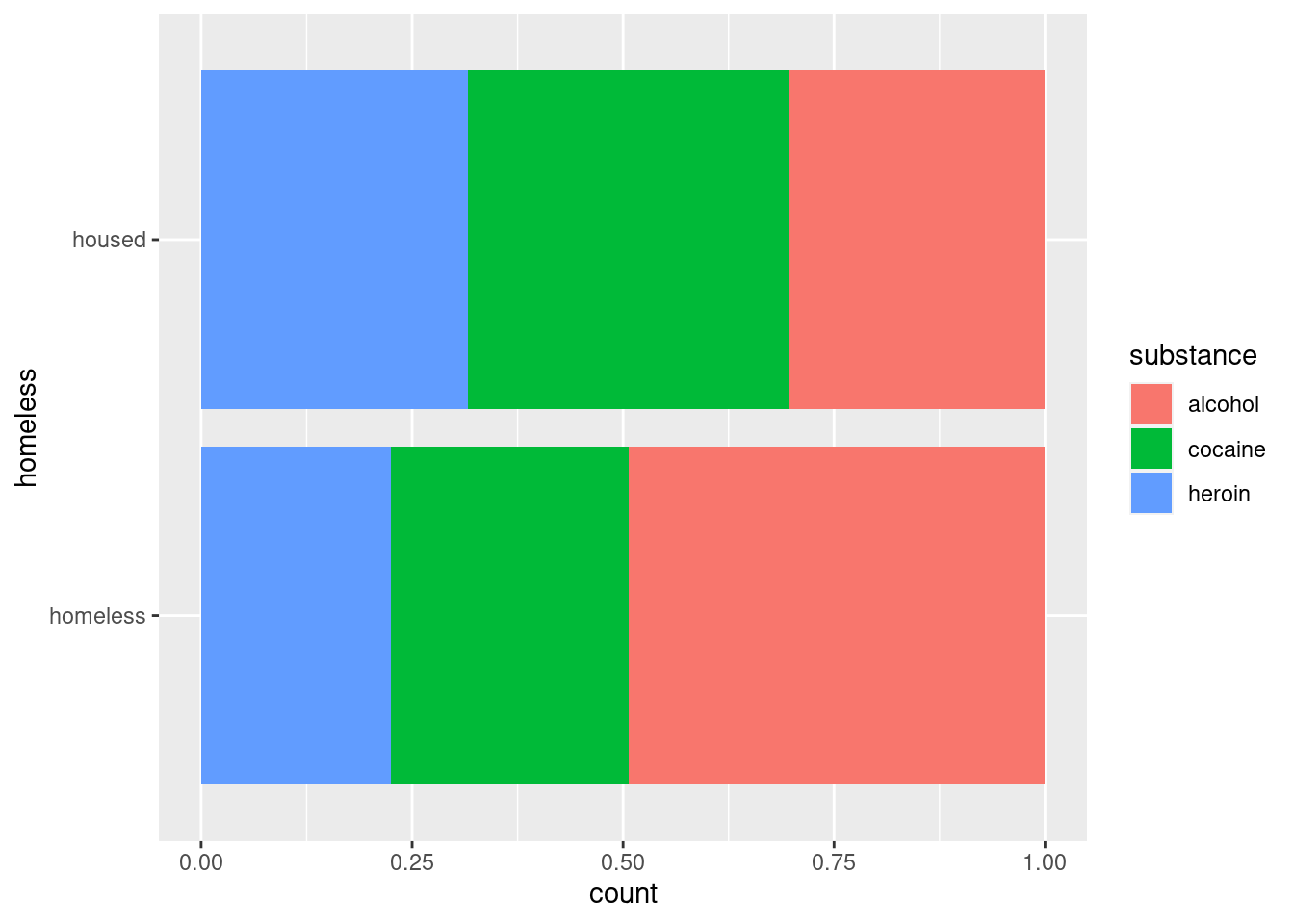
NHANES
Here is the link to the NHANES website.
library(NHANES)
head(NHANES)# A tibble: 6 × 76
ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
<int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
1 51624 2009_10 male 34 " 30-39" 409 White <NA> High School
2 51624 2009_10 male 34 " 30-39" 409 White <NA> High School
3 51624 2009_10 male 34 " 30-39" 409 White <NA> High School
4 51625 2009_10 male 4 " 0-9" 49 Other <NA> <NA>
5 51630 2009_10 female 49 " 40-49" 596 White <NA> Some College
6 51638 2009_10 male 9 " 0-9" 115 White <NA> <NA>
# … with 67 more variables: MaritalStatus <fct>, HHIncome <fct>,
# HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>, HomeOwn <fct>,
# Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>, Height <dbl>,
# BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>, Pulse <int>,
# BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>, BPDia1 <int>, BPSys2 <int>,
# BPDia2 <int>, BPSys3 <int>, BPDia3 <int>, Testosterone <dbl>,
# DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>, UrineFlow1 <dbl>, …Take a sample first and then make the plot.
sample_n(NHANES, size = 1000) %>% ggplot(aes(x = Age, y = Height, color = Gender)) +
geom_point() +
geom_smooth() +
xlab("Age (years)") +
ylab("Height (cm)")`geom_smooth()` using method = 'loess' and formula 'y ~ x'Warning: Removed 31 rows containing non-finite values (stat_smooth).Warning: Removed 31 rows containing missing values (geom_point).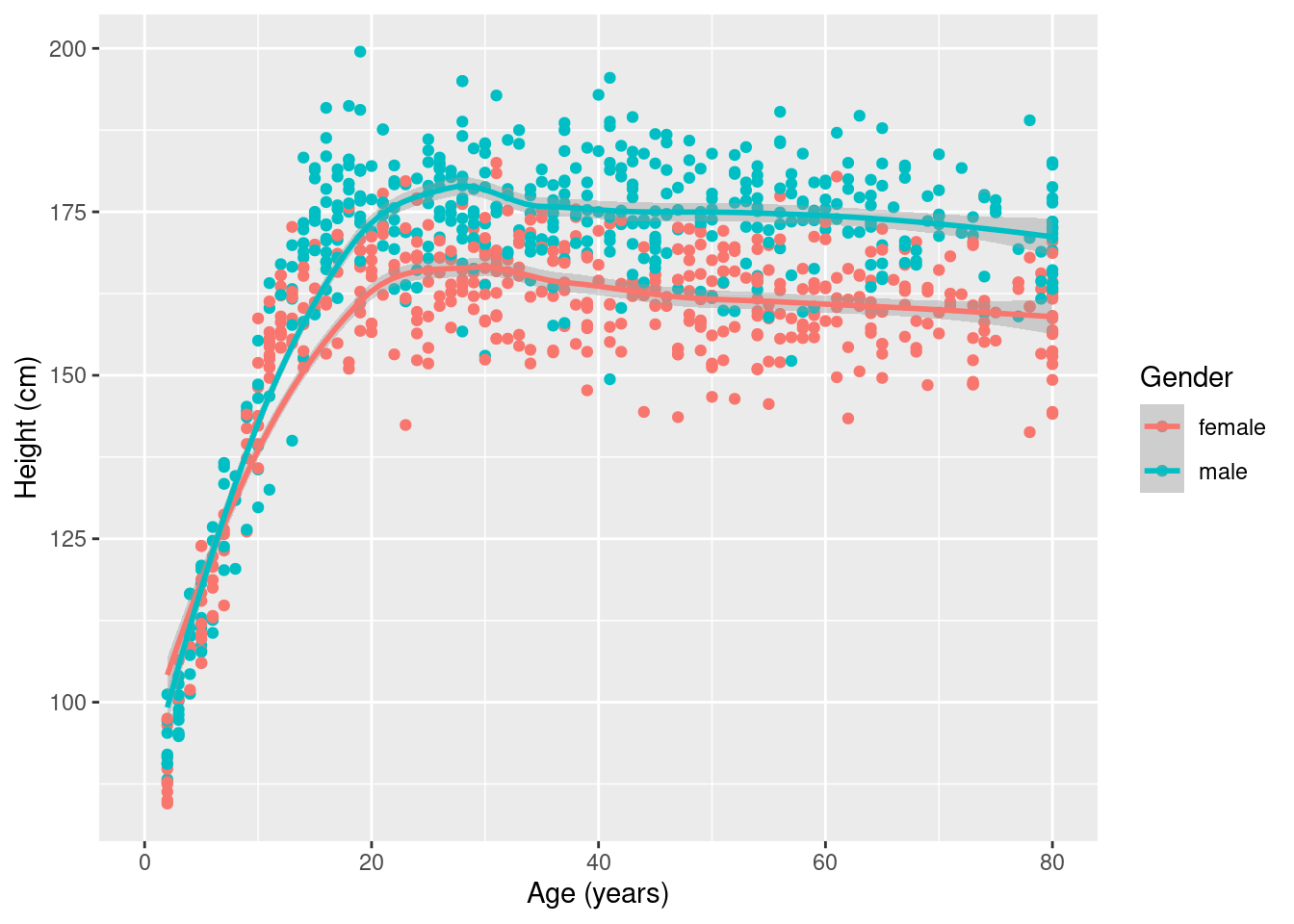
Here is an alternative plot using all the data. This is hexbin plot.
NHANES %>% ggplot(aes(x = Age, y = Height, color = Gender)) +
geom_hex() +
geom_smooth() +
xlab("Age (years)") +
ylab("Height (cm)")Warning: Removed 353 rows containing non-finite values (stat_binhex).`geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'Warning: Removed 353 rows containing non-finite values (stat_smooth).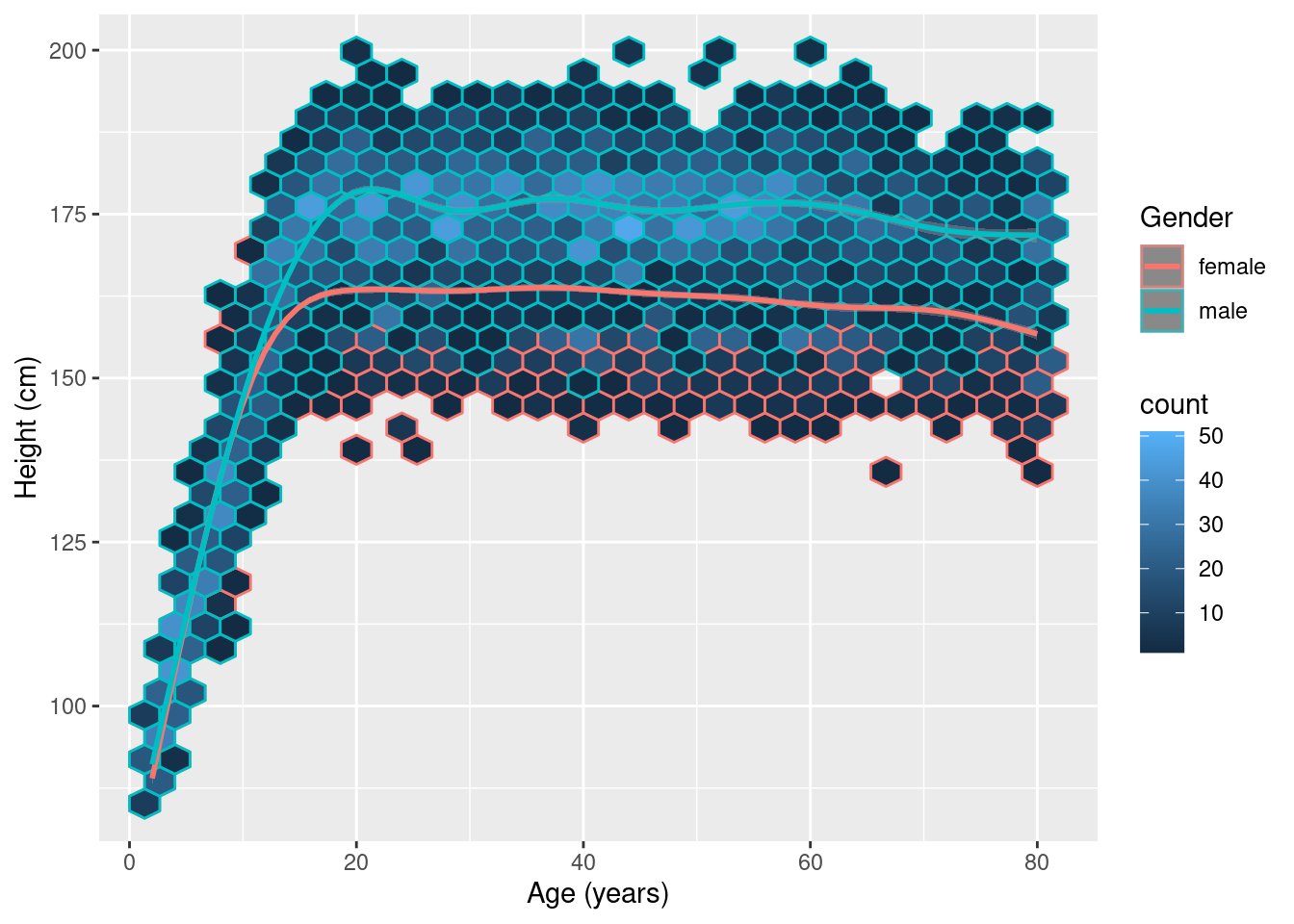
library(mosaic)Registered S3 method overwritten by 'mosaic':
method from
fortify.SpatialPolygonsDataFrame ggplot2
The 'mosaic' package masks several functions from core packages in order to add
additional features. The original behavior of these functions should not be affected by this.
Attaching package: 'mosaic'The following object is masked from 'package:Matrix':
meanThe following objects are masked from 'package:dplyr':
count, do, tallyThe following object is masked from 'package:purrr':
crossThe following object is masked from 'package:ggplot2':
statThe following objects are masked from 'package:stats':
binom.test, cor, cor.test, cov, fivenum, IQR, median, prop.test,
quantile, sd, t.test, varThe following objects are masked from 'package:base':
max, mean, min, prod, range, sample, sumhead(NHANES)# A tibble: 6 × 76
ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
<int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
1 51624 2009_10 male 34 " 30-39" 409 White <NA> High School
2 51624 2009_10 male 34 " 30-39" 409 White <NA> High School
3 51624 2009_10 male 34 " 30-39" 409 White <NA> High School
4 51625 2009_10 male 4 " 0-9" 49 Other <NA> <NA>
5 51630 2009_10 female 49 " 40-49" 596 White <NA> Some College
6 51638 2009_10 male 9 " 0-9" 115 White <NA> <NA>
# … with 67 more variables: MaritalStatus <fct>, HHIncome <fct>,
# HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>, HomeOwn <fct>,
# Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>, Height <dbl>,
# BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>, Pulse <int>,
# BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>, BPDia1 <int>, BPSys2 <int>,
# BPDia2 <int>, BPSys3 <int>, BPDia3 <int>, Testosterone <dbl>,
# DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>, UrineFlow1 <dbl>, …NHANES2 <- NHANES %>% select(AgeDecade, BMI_WHO)
head(NHANES2)# A tibble: 6 × 2
AgeDecade BMI_WHO
<fct> <fct>
1 " 30-39" 30.0_plus
2 " 30-39" 30.0_plus
3 " 30-39" 30.0_plus
4 " 0-9" 12.0_18.5
5 " 40-49" 30.0_plus
6 " 0-9" 12.0_18.5NHANES2_table <- table(NHANES2)
NHANES2_table BMI_WHO
AgeDecade 12.0_18.5 18.5_to_24.9 25.0_to_29.9 30.0_plus
0-9 873 193 28 7
10-19 280 664 244 172
20-29 49 526 349 418
30-39 10 394 433 495
40-49 26 371 475 506
50-59 15 314 487 477
60-69 8 199 321 373
70+ 6 142 207 218mosaicplot(NHANES2_table, color = TRUE)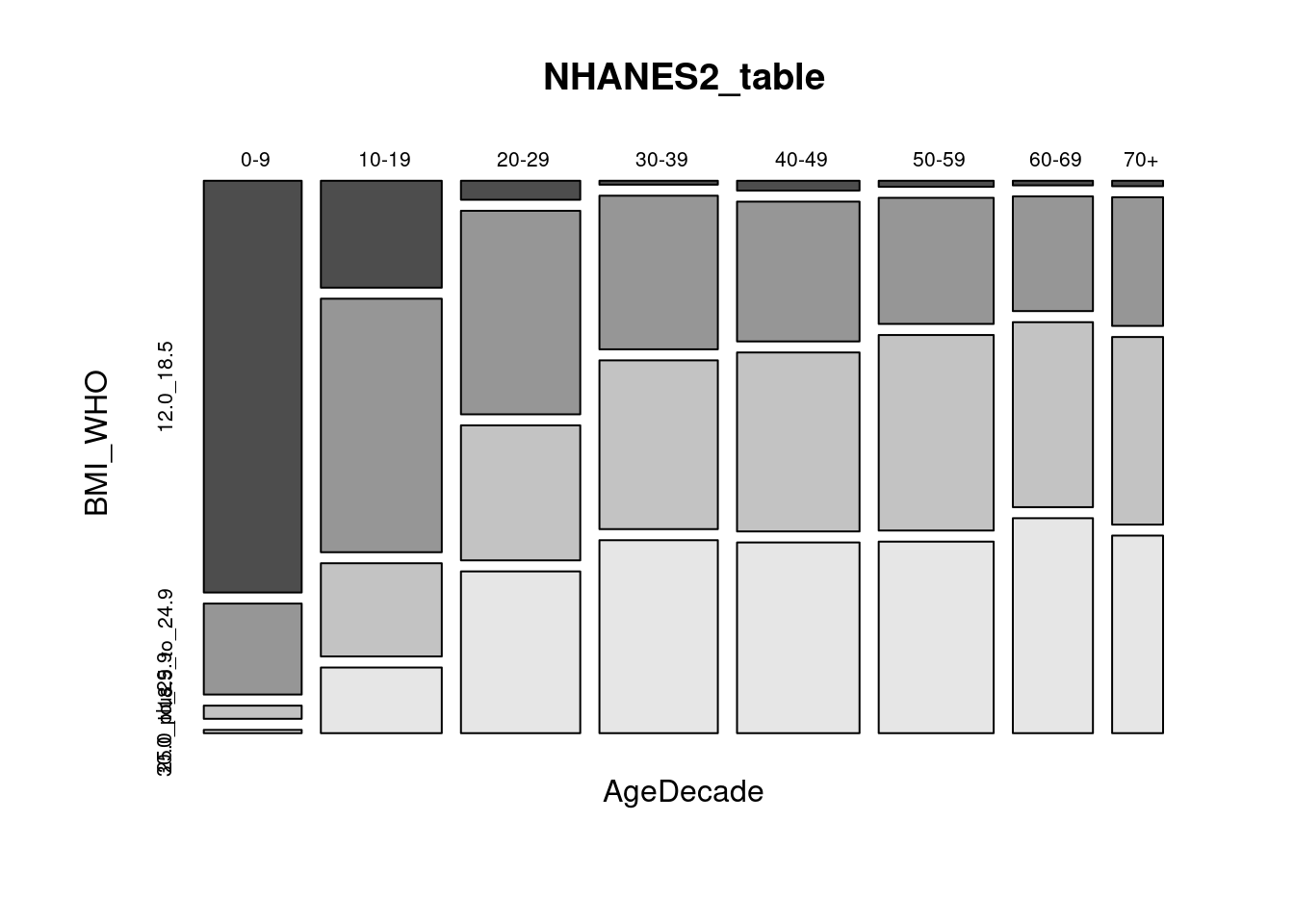
Weather
library(macleish)Loading required package: etlhead(whately_2015)# A tibble: 6 × 8
when temperature wind_speed wind_dir rel_humidity pressure
<dttm> <dbl> <dbl> <dbl> <dbl> <int>
1 2015-01-01 00:00:00 -9.32 1.40 225. 54.6 985
2 2015-01-01 00:10:00 -9.46 1.51 248. 55.4 985
3 2015-01-01 00:20:00 -9.44 1.62 258. 56.2 985
4 2015-01-01 00:30:00 -9.3 1.14 244. 56.4 985
5 2015-01-01 00:40:00 -9.32 1.22 238. 56.9 984
6 2015-01-01 00:50:00 -9.34 1.09 242. 57.2 984
# … with 2 more variables: solar_radiation <dbl>, rainfall <dbl>whately_2015 %>% ggplot(aes(x = when, y=temperature)) +
geom_line(color = "darkgrey") +
geom_smooth() +
xlab(NULL) +
ylab("Tempurature (degrees Fahrenheit)")`geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'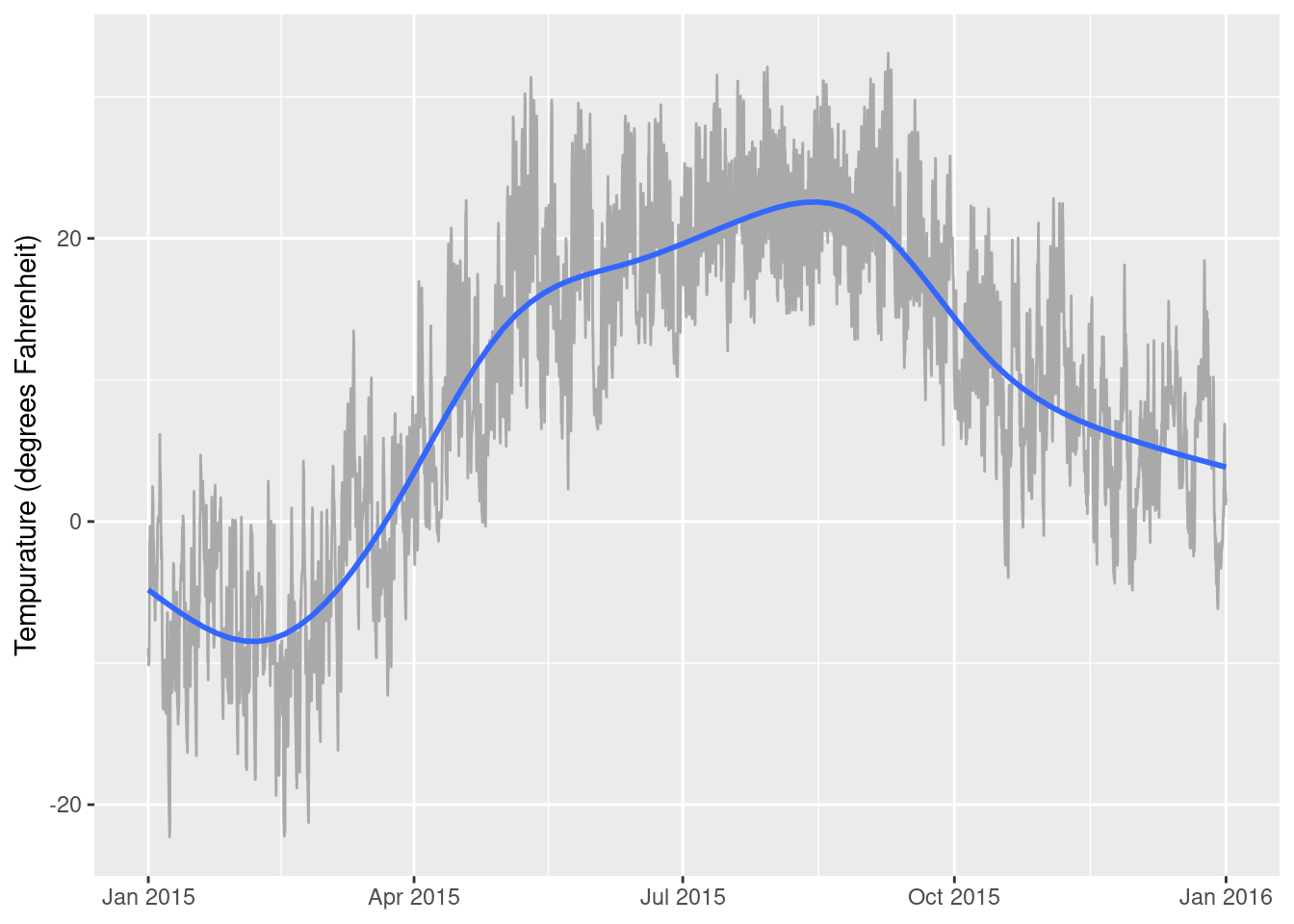
Here is the link to the choroplethr website.
library(choroplethr)Loading required package: acsLoading required package: XML
Attaching package: 'acs'The following object is masked from 'package:dplyr':
combineThe following object is masked from 'package:base':
applylibrary(choroplethrMaps)
data(df_pop_state)
state_choropleth(df_pop_state,
title = "US 2012 State Population Estimates",
legend = "Population")Warning in private$zoom == "alaska" || private$zoom == "hawaii": 'length(x) = 51
> 1' in coercion to 'logical(1)'
Warning in private$zoom == "alaska" || private$zoom == "hawaii": 'length(x) = 51
> 1' in coercion to 'logical(1)'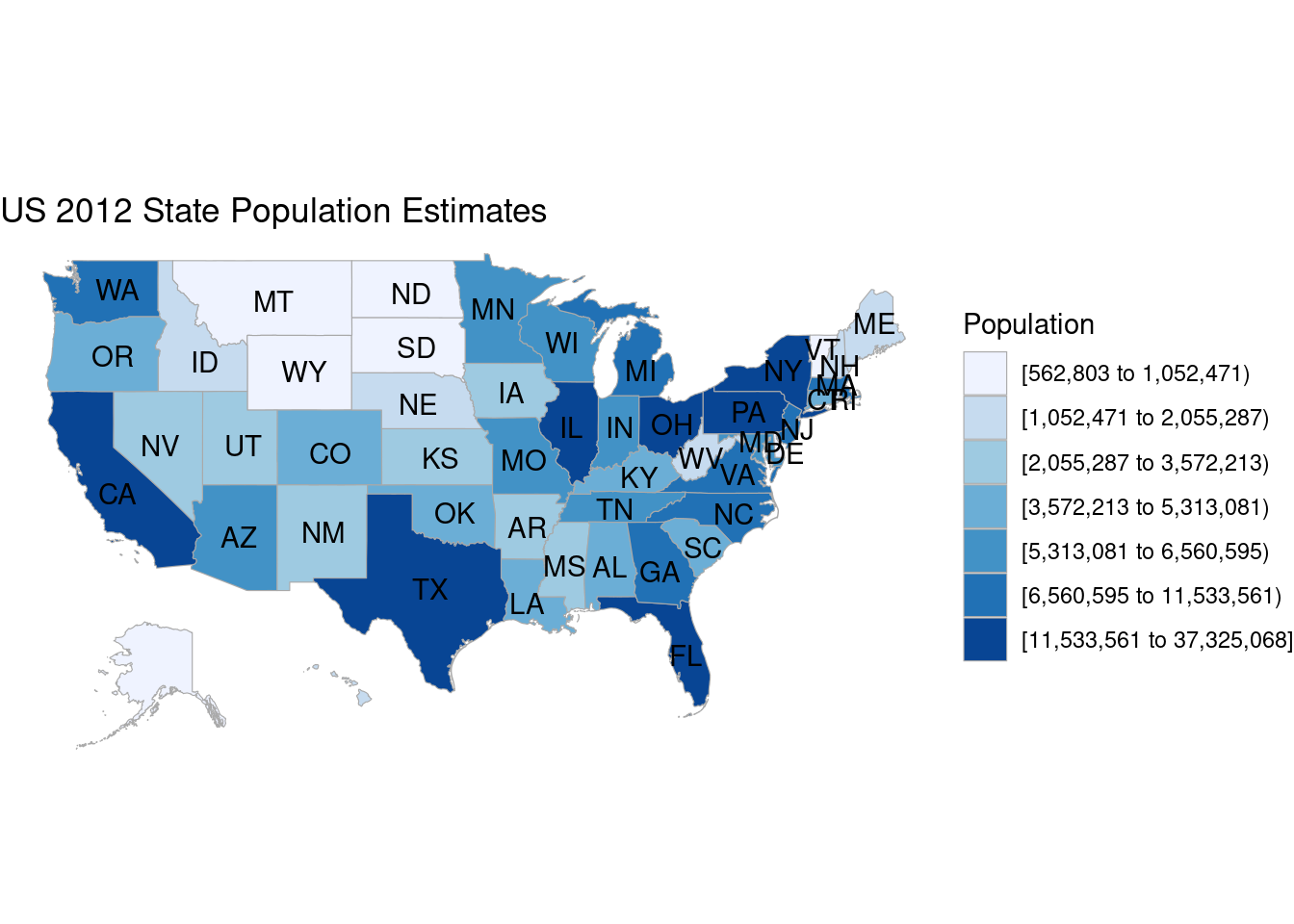
data(df_pop_county)
county_choropleth(df_pop_county,
title = "US 2012 County Population Estimates",
legend = "Population")Warning in private$zoom == "alaska" || private$zoom == "hawaii": 'length(x) = 51
> 1' in coercion to 'logical(1)'
Warning in private$zoom == "alaska" || private$zoom == "hawaii": 'length(x) = 51
> 1' in coercion to 'logical(1)'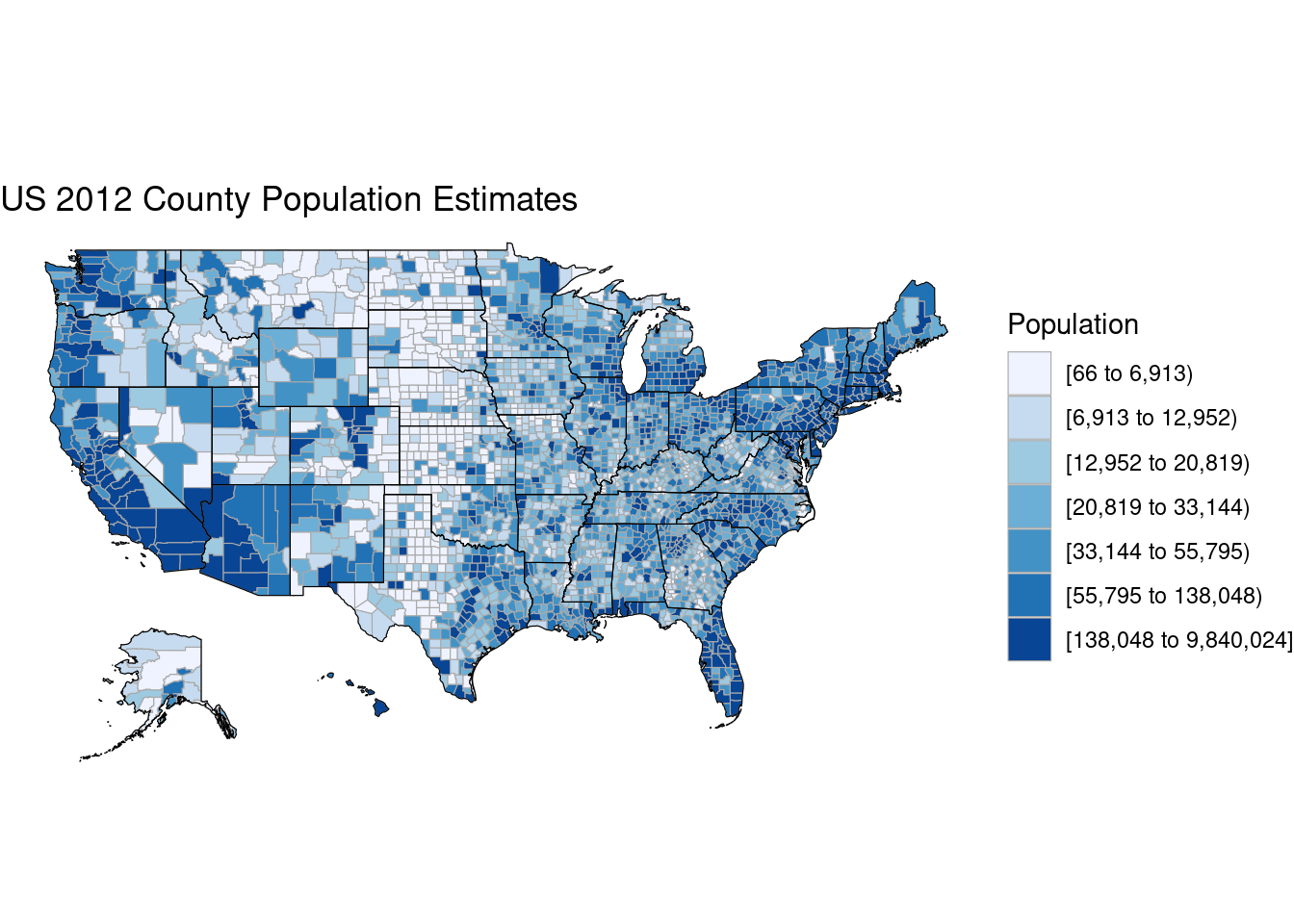
data(df_pop_country)
country_choropleth(df_pop_country, "2012 World Bank Populate Estimates")Warning in self$bind(): The following regions were missing and are being set to
NA: namibia, western sahara, taiwan, antarctica, kosovo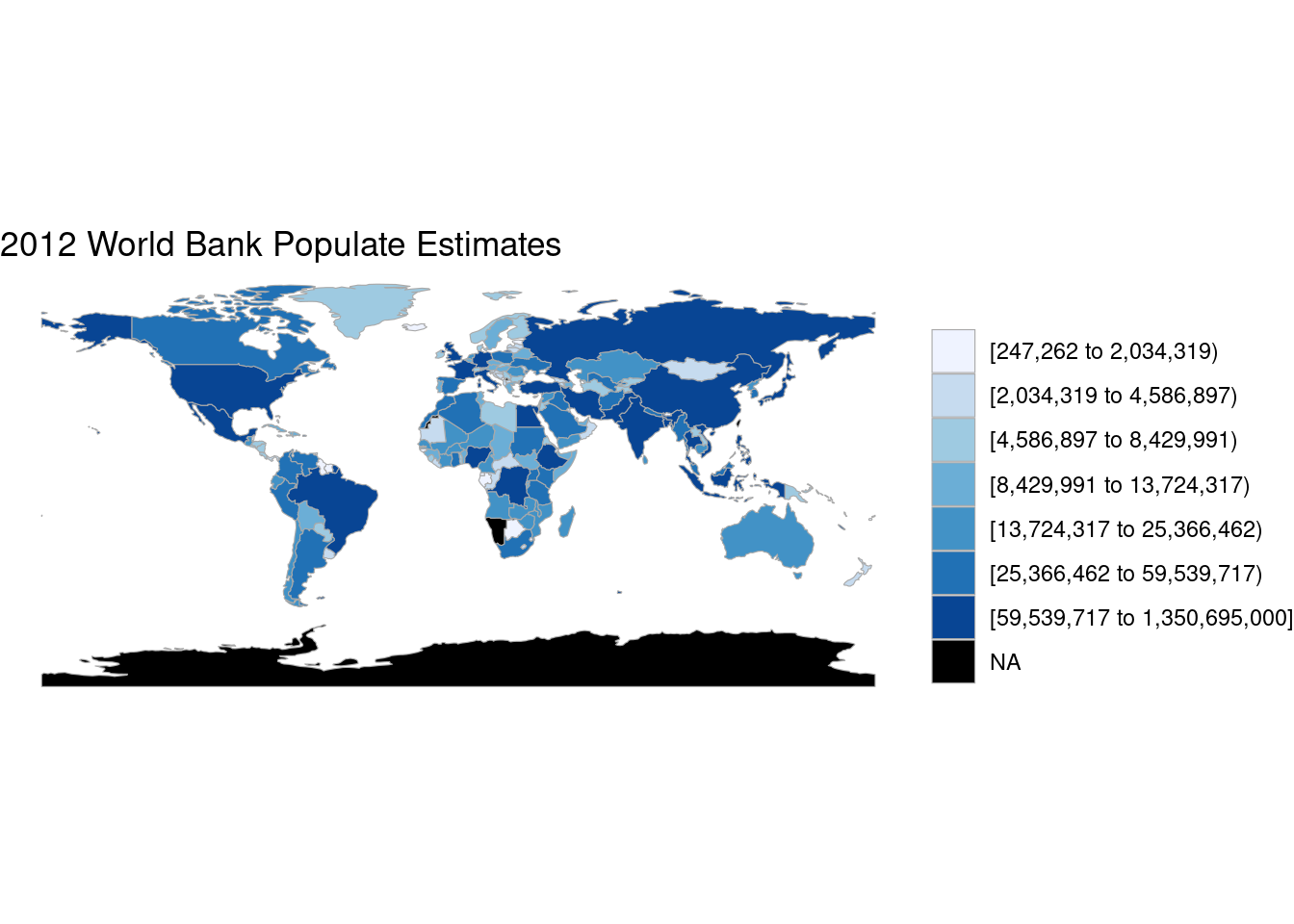
Networks
Check out ggnetwork
Example 4.
library(ggnetwork)
library(network)
'network' 1.17.1 (2021-06-12), part of the Statnet Project
* 'news(package="network")' for changes since last version
* 'citation("network")' for citation information
* 'https://statnet.org' for help, support, and other informationlibrary(sna)Loading required package: statnet.common
Attaching package: 'statnet.common'The following objects are masked from 'package:base':
attr, ordersna: Tools for Social Network Analysis
Version 2.6 created on 2020-10-5.
copyright (c) 2005, Carter T. Butts, University of California-Irvine
For citation information, type citation("sna").
Type help(package="sna") to get started.n <- network(rgraph(10, tprob = 0.2), directed = FALSE)
n %v% "family" <- sample(letters[1:3], 10, replace = TRUE)
n %v% "importance" <- sample(1:3, 10, replace = TRUE)
e <- network.edgecount(n)
set.edge.attribute(n, "type", sample(letters[24:26], e, replace = TRUE))
set.edge.attribute(n, "day", sample(1:3, e, replace = TRUE))
ggnetwork(n, layout = "fruchtermanreingold", cell.jitter = 0.75) x y family importance vertex.names xend yend day
1 0.0000000 0.3591656 a 2 7 0.1790696 0.5625896 2
3 0.3402737 0.6676026 b 3 5 0.3990675 0.2671296 3
4 0.3436387 0.4217009 c 1 8 0.6730268 0.3301077 3
5 0.3436387 0.4217009 c 1 8 0.3990675 0.2671296 2
6 0.3436387 0.4217009 c 1 8 0.0000000 0.3591656 1
7 0.3436387 0.4217009 c 1 8 0.3402737 0.6676026 1
8 0.3436387 0.4217009 c 1 8 0.5380735 0.6840422 2
9 0.3990675 0.2671296 a 1 4 0.1790696 0.5625896 1
10 0.3990675 0.2671296 a 1 4 0.6730268 0.3301077 1
11 0.5380735 0.6840422 a 1 6 0.1790696 0.5625896 1
12 0.5380735 0.6840422 a 1 6 0.3402737 0.6676026 1
13 0.5380735 0.6840422 a 1 6 0.6730268 0.3301077 1
14 0.5988985 0.0000000 a 3 9 0.3990675 0.2671296 2
15 0.5988985 0.0000000 a 3 9 0.6730268 0.3301077 1
16 0.6730268 0.3301077 c 2 3 1.0000000 0.2853717 3
17 0.7075406 1.0000000 b 1 10 0.5380735 0.6840422 2
18 1.0000000 0.2853717 a 3 1 1.0000000 0.2853717 NA
2 0.1790696 0.5625896 c 2 2 0.1790696 0.5625896 NA
31 0.6730268 0.3301077 c 2 3 0.6730268 0.3301077 NA
41 0.3990675 0.2671296 a 1 4 0.3990675 0.2671296 NA
51 0.3402737 0.6676026 b 3 5 0.3402737 0.6676026 NA
61 0.5380735 0.6840422 a 1 6 0.5380735 0.6840422 NA
71 0.0000000 0.3591656 a 2 7 0.0000000 0.3591656 NA
81 0.3436387 0.4217009 c 1 8 0.3436387 0.4217009 NA
91 0.5988985 0.0000000 a 3 9 0.5988985 0.0000000 NA
101 0.7075406 1.0000000 b 1 10 0.7075406 1.0000000 NA
type
1 y
3 x
4 z
5 x
6 x
7 z
8 y
9 x
10 z
11 z
12 y
13 x
14 x
15 x
16 z
17 y
18 <NA>
2 <NA>
31 <NA>
41 <NA>
51 <NA>
61 <NA>
71 <NA>
81 <NA>
91 <NA>
101 <NA>ggnetwork(n, layout = "target", niter = 100) x y family importance vertex.names xend yend
1 0.00000000 0.46498651 a 3 9 0.63884925 0.44298698
2 0.00000000 0.46498651 a 3 9 0.60812206 0.41357881
4 0.24558221 1.00000000 b 1 10 0.59395584 0.47774812
5 0.59395584 0.47774812 a 1 6 0.95382151 0.28021317
6 0.59395584 0.47774812 a 1 6 0.95933695 0.60247559
7 0.59395584 0.47774812 a 1 6 0.60812206 0.41357881
8 0.60812206 0.41357881 c 2 3 0.05122724 0.00000000
9 0.62449835 0.47004012 c 1 8 0.60812206 0.41357881
10 0.62449835 0.47004012 c 1 8 0.63884925 0.44298698
11 0.62449835 0.47004012 c 1 8 1.00000000 0.09447686
12 0.62449835 0.47004012 c 1 8 0.95933695 0.60247559
13 0.62449835 0.47004012 c 1 8 0.59395584 0.47774812
14 0.63884925 0.44298698 a 1 4 0.95382151 0.28021317
15 0.63884925 0.44298698 a 1 4 0.60812206 0.41357881
17 0.95933695 0.60247559 b 3 5 0.63884925 0.44298698
18 1.00000000 0.09447686 a 2 7 0.95382151 0.28021317
16 0.05122724 0.00000000 a 3 1 0.05122724 0.00000000
21 0.95382151 0.28021317 c 2 2 0.95382151 0.28021317
3 0.60812206 0.41357881 c 2 3 0.60812206 0.41357881
41 0.63884925 0.44298698 a 1 4 0.63884925 0.44298698
51 0.95933695 0.60247559 b 3 5 0.95933695 0.60247559
61 0.59395584 0.47774812 a 1 6 0.59395584 0.47774812
71 1.00000000 0.09447686 a 2 7 1.00000000 0.09447686
81 0.62449835 0.47004012 c 1 8 0.62449835 0.47004012
91 0.00000000 0.46498651 a 3 9 0.00000000 0.46498651
101 0.24558221 1.00000000 b 1 10 0.24558221 1.00000000
day type
1 2 x
2 1 x
4 2 y
5 1 z
6 1 y
7 1 x
8 3 z
9 3 z
10 2 x
11 1 x
12 1 z
13 2 y
14 1 x
15 1 z
17 3 x
18 2 y
16 NA <NA>
21 NA <NA>
3 NA <NA>
41 NA <NA>
51 NA <NA>
61 NA <NA>
71 NA <NA>
81 NA <NA>
91 NA <NA>
101 NA <NA>head(ggnetwork(n)) x y family importance vertex.names xend yend day
2 0.3269990 0.7049237 c 2 3 0.0000000 0.7662902 3
3 0.4028999 1.0000000 a 3 9 0.5338945 0.6220794 2
4 0.4028999 1.0000000 a 3 9 0.3269990 0.7049237 1
5 0.4592103 0.0000000 a 2 7 0.6584518 0.2469472 2
6 0.4720168 0.3626698 c 1 8 0.3269990 0.7049237 3
7 0.4720168 0.3626698 c 1 8 0.5338945 0.6220794 2
type
2 z
3 x
4 x
5 y
6 z
7 xtail(ggnetwork(n)) x y family importance vertex.names xend yend day
51 0.1104049 0.5085459 b 3 5 0.1104049 0.5085459 NA
61 0.3178598 0.3809046 a 1 6 0.3178598 0.3809046 NA
71 0.0000000 1.0000000 a 2 7 0.0000000 1.0000000 NA
81 0.2818331 0.7101530 c 1 8 0.2818331 0.7101530 NA
91 0.7790771 0.9340538 a 3 9 0.7790771 0.9340538 NA
101 0.3136755 0.0000000 b 1 10 0.3136755 0.0000000 NA
type
51 <NA>
61 <NA>
71 <NA>
81 <NA>
91 <NA>
101 <NA>ggplot(n)
ggplot(ggnetwork(n))
ggplot(n, aes(x = x, y = y, xend = xend, yend = yend)) +
geom_edges(aes(linetype = type), color = "grey50") +
theme_blank()
ggplot(n, aes(x = x, y = y, xend = xend, yend = yend)) +
geom_edges(aes(linetype = type), color = "grey50", curvature = 0.1) +
theme_blank()
ggplot(n, aes(x = x, y = y, xend = xend, yend = yend)) +
geom_edges(aes(linetype = type), color = "grey50") +
geom_nodes(aes(color = family, size = importance)) +
theme_blank()
ggplot(n, aes(x = x, y = y, xend = xend, yend = yend)) +
geom_edges(color = "black") +
geom_nodes(color = "black", size = 8) +
geom_nodetext(aes(color = family, label = LETTERS[ vertex.names ]),
fontface = "bold") +
theme_blank()
Review Table 3.3 on page 47 for the different kinds of plots that can be made for different kinds of x, y variables.
Continue with the Extended example: Historical baby names on page 48.